INTRODUCTION
C. ebano H. Karst. (Fabaceae: Caesalpinioideae) is a tropical tree from Colombia that is distributed throughout the country. The members of this subfamily often present a mixed pollination system since they can self-pollinate or have cross-pollination and can be found in mountainous areas of the tropical dry forest (bs-T). Their conservation status is in the endangered category (EN) given the few populations that remain today (Fremout et al., 2021). Under a technical context, its wood is black and of high quality, so its demand was high, intending to export and construct architectural structures; today, it is threatened by its use as firewood and charcoal (Palencia et al., 2017; Castro-Torres et al., 2021).
Ebony has potential in urban forestry (silviculture) given its medium growth, high wood density, ability to capture carbon, adaptability to urban environments, and aesthetic appeal; it is also recommended for sites without height restrictions or in partnership with agroforestry systems aimed at ecological restoration and conservation of the bs-T (Arenas-Rubio, 2019). Recent research has evaluated the effect of the addition of organic amendments and microbial inoculants on nursery growth (Hernández-Gómez et al., 2018), forest production for potential use in agroforestry systems (Arenas-Rubio, 2019), and its diversity and genetic structure as a characteristic species of bs-T (Palencia et al., 2017; Bocanegra-González et al., 2019; Castro-Torres et al., 2021).
In genetic improvement programs, correlations between traits of agronomic and silvicultural interest are important to facilitate and increase selection efficiency (Espitia et al., 2017). The improvement of a trait of interest can be done through the indirect selection of other traits that are associated with it and less complex, with higher heritability, which would make the selection more efficient and safe (Gupta et al., 2017; Araméndiz-Tatis et al., 2020; Araméndiz-Tatis 2021). Studies of associations between traits based on their phenotypic and genetic correlations make it possible to improve the efficiency of selection in genetic improvement programs, improve the quality of the products, and achieve greater genetic progress (Pérez-Pelea et al., 2018; Araméndiz-Tatis et al., 2020; Araméndiz-Tatis et al., 2021). Several authors have developed correlation studies between the biometric traits of the fruit and its seeds in forest species (Leão et al., 2017; Dos-Santos et al., 2018; Ristau et al., 2018).
According to Gupta et al. (2017) some traits are often influenced by variations in other traits, so care must be taken with the simple interpretation of the correlation value since direct selection ends up being inefficient (Araméndiz-Tatis et al., 2020). In this sense, Lekshmanan & Vahab (2018) pointed out that the statistical analysis of the path is proposed as a solution to decompose the correlations into direct effects of X on Y and the indirect effects of X on Y via other independent variables in a correlation system. The path coefficients are standardized regression coefficients that split the estimated correlations into their components: the direct effect for the predictor variable and the indirect effects on the dependent character variations (Lekshmanan & Vahab, 2018; Agrawal et al., 2018).
Studies on phenotypic and genetic correlations and path analysis in forest trees based on fruit and seed characters are not very prolific (Espitia et al., 2018). This situation is more serious in C. ebano given that in Colombia, there is not much research that contributes to a much safer selection process in the genetic advancement of the species. It is possibly because there are no reports of natural or planted populations being isolated and dispersed individuals in urban, peri-urban, or rural areas, which have originated as a result of the dispersion of their seeds under the anthropic or natural influence.
This study aimed to evaluate the correlations and path analysis between the biometric traits of the fruit and seeds of C. ebano in Montería (Córdoba, Colombia) and to provide information for future programs of genetic improvement, sustainable conservation, and rational use of forest germplasm in the country.
MATERIALS AND METHODS
Location and time of evaluation. The field study was carried out in 2018 on fruits harvested at random from 10 C. ebano trees located at the University of Córdoba in Montería (Córdoba - Colombia). The fruits and their seeds were processed in the Plant Breeding Laboratory of the University of Córdoba, located in the same city, at 8°47'31.9"N - 75° 51'38.6"W. The place is located at an altitude of 15 m and belongs to the tropical dry forest life zone (bs-T) of the middle Sinú Valley, with an average temperature of 28.0°C, relative humidity of 84%, and annual rainfall of 1200mm (Castro-Torres et al., 2021).
Genetic material. In this study, 10 C. ebano trees taken from the arboretum of the University of Córdoba were evaluated, from which random samples of 10 fruits per tree were taken, for a total of 100 fruits. Due to allogamous pollination, the seed coming from each tree was considered a half-sibling family. The trees sampled at the time of fruit collection were between 6 and 15 years old, between 5 and 10m tall, and 1.3m in diameter, between 25 and 40cm.
Response variables. Nine biometric traits of the fruit and seeds were measured: fresh fruit weight (FFW), fruit length (FL), fruit width (FW), fruit thickness (FT), number of seeds per fruit (NSF), total weight of seeds per fruit (TWSF), fresh weight of a seed (W1S), volume of a seed (V1S) and density of a seed (D1S).
The FFW and TWS were determined with a precision balance BJ410C in grams; FL, FW, and FT were measured with a flexometer and vernier in centimeters; NSF was determined by counting the seeds after extracting them from the fruit; W1S was obtained as the average weight, in grams, of all the seeds of each fruit (TWSF/NSF); the V1S, as the increase in volume generated in a test tube, with known distilled water volume, when introducing four seeds taken at random from each fruit, and the D1S was calculated from the W1S/V1S.
Statistical analysis. The 10 trees were evaluated assuming the random-effects model for the analysis of variance, with 10 treatments (trees) and 10 replications (fruits/tree). The analyses of variance and covariance for the response variables: FFW, FL, FW, FT, NSF, TWSF, W1S, V1S, and D1S, the phenotypic and genetic correlations, and path analysis for the FFW were performed using the freely accessible computer program GENES version Windows GENES V.2016.6.0 (Cruz, 2016). The GENES program applies the classic correlation, Equation 1, Equation 2:
Where: r(xy) and COV(xy) are the phenotypic and genetic correlations and covariances between traits X and Y, respectively;/are the phenotypic and genetic variances of X and Y, respectively.
Two path analyses were performed for the system: FFW (effect variable: Y) based on FL (X1), FW (X2), FT (X3), NSF (X4), TWSF (X5), W1S (X6), V1S (X7), and D1S (X8) (cause variables). These two analyses originated from the phenotypic and genetic correlation matrices, previously generated automatically by the GENES program in the analysis of variance. The direct effects in each of the analyses are estimated by breaking down the phenotypic and/or genetic correlation matrix into the following matrix system: P = A-1*R, where: A-1 is the inverse of the correlation matrix (between each of the cause variables), R is the correlation coefficient vector between the cause variables (Xi) with the effect variable (Y), and P is the path coefficients vector (Espitia et al., 2018).
The decomposition of the correlations of each of the cause variables with the effect variable (rXiYi = 1,2,3, ..., 8) into its direct effect (Pi) and indirect effect (Ei) components allows estimating the respective indirect effects (Ei) of each cause variable by solving the following equations (Espitia et al., 2018): rx1y = P1 + E1: for the FL; rx2y = P2 + E2: for the FW; rx3y = P3 + E3: for the FT; rx4y = P4 + E4: for the NSF; rx5y = P5 + E5: for the TWSF; rx6y = P6 + E6: for the W1S; rx7y = P7 + E7: for the V1S yrx8y = P8 + E8: for D1S.
The path coefficient due to residual effects or other traits not considered in the study (h) was estimated using the following Equation 3:
RESULTS AND DISCUSSION
Analysis of variance. The mean squares for the analysis of variance, their statistical significance, and the general mean for the nine variables considered in the study are presented in Table 1. Highly significant variability (p<0.01) was detected for fresh fruit weight (FFW), fruit length (FL), fruit thickness (FT), number of seeds per fruit (NSF), total weight of seeds per fruit (TWSF), volume (V1S), and density of a seed (D1S) (Table 1). Significant variations in biometric traits among ebony trees in the Colombian Caribbean suggest the existence of genetic variability, allowing genetic progress to be achieved in plant breeding programs through the selection of trees with desirable fruit and seed characters (Dos-Santos et al., 2018; Espitia et al., 2018). Similar results were reported in forest species of the same ebony family by Okello et al. (2018) in Tamarindus indica L. and other economically important species such as Hymenae acourbaril L. (Ristau et al., 2018), Bursera simplex Rzed. & Calderón (Luna et al., 2018) and Pachira aquatica Aubl. (Espitia et al., 2018).
Table 1 Mean squares and statistical significance of the analysis of variance for nine biometric traits of the fruit and seeds of C. ebano H. Karst trees.
| Trait1 | CMA | CME | Mean | CV (%) |
| FFW | 16.13343** | 2.767967 | 7.96 | 25.06 |
| FL | 6.237333** | 0.978267 | 7.64 | 12.94 |
| FW | 0.109511ns | 0.061044 | 2.70 | 9.14 |
| FT | 0.1624** | 0.050533 | 0.95 | 23.61 |
| NSF | 16.356667** | 4.423333 | 4.63 | 45.71 |
| TWSF | 0.155534** | 0.038683 | 0.44 | 45.02 |
| W1S | 0.000132ns | 0.000162 | 0.10 | 13.26 |
| V1S | 0.000601** | 0.000202 | 0.08 | 17.56 |
| D1S | 0.135557** | 0.051248 | 1.22 | 18.53 |
1FFW: fresh fruit weight; FL: fruit length; FW: fruit width; FT: fruit thickness; NSF: number of seeds per fruit; TWSF: total weight of seeds per fruit; W1S: weight of a seed; V1S: volume of a seed; D1S: density of a C. ebano H. Karst seed; CMA: mean square between trees; CME: experimental error (mean squares within); CV = coefficient of variation; ns: not significant; *: p<0.01
The high variability for these phenotypic characters is a response to the edaphoclimatic and genetic variations, expressed differentially between trees (Luna et al., 2018). Preliminary studies reported similar results at the level of fruit and seed characteristics, physiological parameters of germination, and seedling development in forest species (Dos-Santos et al., 2018; Okello et al., 2018). Other factors at the level of individuals and evaluated characteristics, such as type of population and pollination, geographical position, nutrition and age of the mother plant, deterioration and level of maturity at harvest, and the genotype environment interaction, can also explain the differences detected between the values (Luna et al., 2018). The absence of significance for FW and W1S suggests a close common genetic origin or high genetic kinship due to possible effects of inbreeding in isolated individuals, which is a fact that reduces the possibility of achieving genetic advancement using these characteristics (Sujii et al., 2017; Espitia et al., 2018).
Phenotypic and genetic correlations. The phenotypic (rF's) and genetic (rG's) correlation coefficients between the nine biometric characters of fruits and seeds studied in C. ebano are presented in Table 2. From the 64 possible correlations of the study, only 19 phenotypic (29.7%) and 23 genetic (35.9%) correlations were found to be significant (p<0.05 and p<0.01) and positive or negative. It was observed that, among the biometric characters of the fruit, the weight of the fruit (FFW) presented high significant correlations (p<0.01) and positive with the length of the fruit (FL) (rF≥0.98** and rG≥1.00**) and with the width of the fruit (FW) (rF=0.80** and rG=1.00**); a similar situation occurred with the number of seeds per fruit (NSF) and the total weight of seeds per fruit (TWSF), which had highly significant association at the phenotypic and genetic level (rF≥0.89** and rG≥0.94**, respectively); while the degree of association with seed density (D1S) was significant (rF=0.59* and rG=0.68*).
Table 2 Phenotypic (rF) and genetic (rG) correlations between nine biometric traits of the fruit and seeds of C. ebano H. Karst trees.
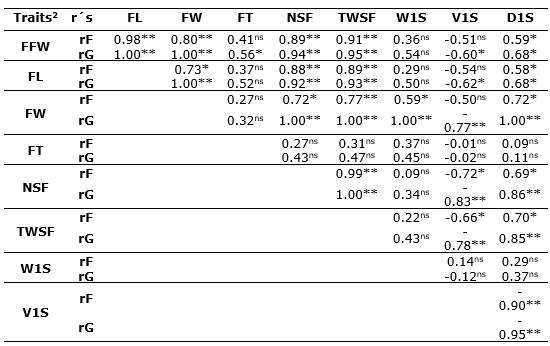
2FFW: fresh fruit weight; FL: fruit lengtht; FW: fruit width; FT: fruit thickness; NSF: number of seeds per fruit; TWSF: total weight of seeds per fruit; W1S: weight of a seed; V1S: volume of a seed; D1S: density of a seed. ns: not significant; *: p <0.05, **: p <0.01.
FL was associated both phenotypically and genetically (p<0.05) with FW, NSF, TWSF, and D1S (rF≥0.58* and rG≥0.68*). A similar situation with higher genetic correlation levels was observed for fruit width (FW), which showed important phenotypic (rF≥0.59*) and perfect genetic correlations (rG=1.00**) with NSF, TWSF, W1S, and D1S; while with V1S the relationship was high and significant, but in the opposite direction (rG=-0.77**). NSF was positive and highly associated with TWSF and D1S (rF≥0.69* and rG≥0.86**); while the relationship with V1S was high and significant but negative (rG=-0.83**).
The TWSF showed a similar association to the NSF with the other variables, presenting significant and positive genetic correlations with FFW, FL, FW, NSF, and D1S (rG≥0.85**) and negative with V1S (rG=-0.78**). The W1S only showed a high, positive, and significant genetic correlation with FW (rG=1.00**), possibly because they are traits controlled by genes with pleiotropic or linked effects, which from the physiological point of view allows the plant to produce photoassimilates that increase FW (Tirkey et al., 2022). The volume of a seed (V1S) presented a significant and negative genetic correlation with all study variables (rG≥-0.60*), except for FT and W1S. The trait D1S was positively and significantly associated (rF's≥0.58* and rG's≥0.68*) with FFW, FL, FW, NSF, and TWSF, and negatively associated with V1S (rF=-0.90** and rG=-0.95**) (Table 2).
Without exception, genetic correlations were found to be greater in magnitude and statistical significance than phenotypic correlations. To the plant breeder, genetic correlations are more important when it is desired to improve populations, and these correlations can be supported by the pleiotropic action of a gene or due to gene linkage. Conversely, phenotypic correlations are of great risk and little practical value since they can lead to errors due to environmental instability (Espitia et al., 2017; Espitia et al., 2018; Schober et al., 2018; Pérez-Pelea et al., 2018).
The estimated positive and significant genetic correlations allow us to deduce, according to the authors Bandi et al. (2018), that the control of associated traits is given by a gene with pleiotropic effects or by several linked genes. In this case, the former is more likely since the characteristics represent the growth of similar tissues in the tree. Afonso et al. (2017) and Zuffo et al. (2019) determined similar associations between fruit and seed traits in Ceiba speciosa (A.St.-Hil.) Ravenna and Hancornia speciosa Gomes, respectively. They also mentioned that selecting fruits of greater size and weight can lead to improvements in other important traits, such as the number and weight of seeds per fruit in native Brazilian tree species.
The trait that presented the greatest magnitude of negative rG's was the V1S (-0.60*≥rG's≤-0.95**), which showed that an increase in any of the traits related to this would lead to a detriment of the volume of the seeds and vice versa. This contrasts with Espitia et al. (2018) in P. aquatica, where V1S was positively correlated with most of the variables evaluated. The absence of correlation and the lack of significance for the remaining 34.4% of the traits (Table 2) is possibly due to some indirect masking effects that other traits have on the real magnitude and meaning of the correlation between them.
The estimated genetic correlation between the FFW, FL, and FW (rG=1.00**) (Table 2) is of great importance in the processes of selection, management, and conservation of C. ebano seeds because it is possible to make a selection of trees with high NSF, TWSF, and D1S indirectly through greater FFW, FL, and/or FW; they are much easier, faster, and cheaper characters to estimate compared to the NSF and TWSF. This could facilitate the selection process, which would be done for any of the correlated traits, according to other studies (Leão et al., 2017; Okello et al., 2018). On the other hand, the selection of fruits with a higher TWSF will contribute to increase in fresh fruit weight and possibly will improve the physiological quality of seeds (Zuffo et al., 2019).
Path analysis for fruit weight. The decomposition of phenotypic and genetic correlations for the average fresh fruit weight (FFW) of C. ebano, in its direct (diagonal in bold) and indirect (rows without bold) effects, are shown in Table 3. The estimates of the coefficients of determination (R2) and the residual effects (h) for the model of phenotypic (rF's) and genotypic (rG's) matrices indicate a good fit of the model for the studied characters since they explained more than 99% of the behavior of the FFW (variable effect) and low residuality (< 0.083).
Table 3 Path analysis for the phenotypic (rF) and genetic (rG) correlations of fruit weight (FFW) of C. ebano H. Karst., based on eight biometric traits of the fruit and seeds.
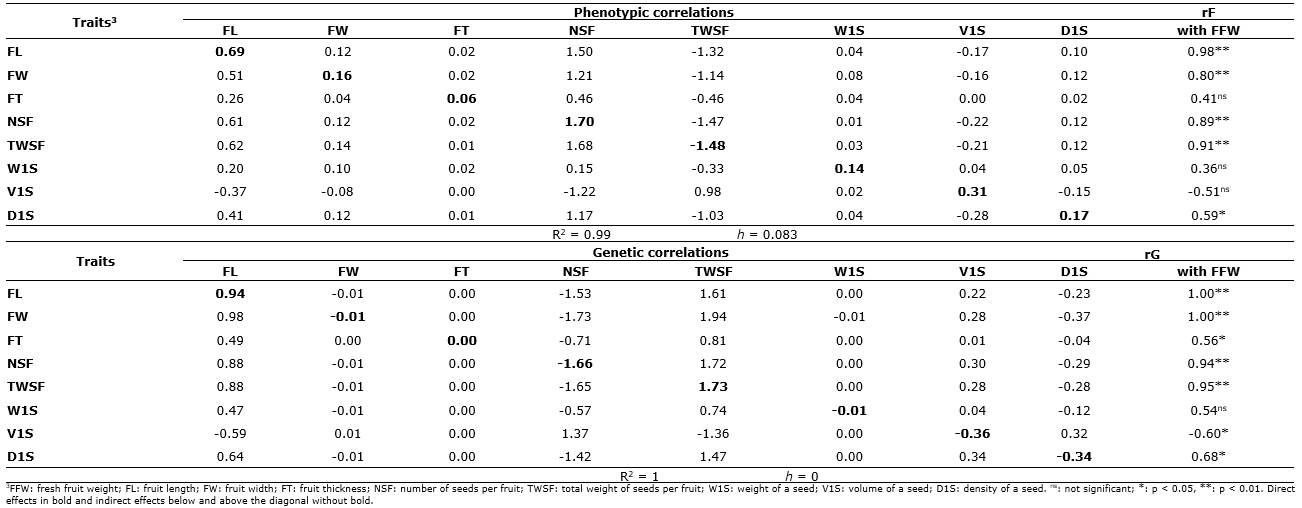
In path analysis using phenotypic correlations (rF's), the traits NSF (PF=1.70), TWSF (PF=-1.48) and FL (PF=0.69) presented the highest direct effects or path coefficients. This explains the degree of association between these variables and the FFW, which showed the highest magnitudes (rF=0.89** to 0.98**).
Path analysis with the use of genetic correlations (rG's) for the FFW indicates that the characteristics that showed the greatest direct effects were the TWSF (PG=1.73), NSF (PG=-1.66), and FL (PG = 0.94). These traits coincide in their behavior with the path analysis based on phenotypic correlations. The genotypic correlation between FL and FFW (rG=1.00**) was explained in greater proportion via indirect effect through NSF (-1.53) and TWSF (1.61); the magnitude and meaning of the association between FW and FFW (rG= 1.00**) were explained via indirect effect through the variables NSF (-1.73) and TWSF (1.94). On the other hand, the magnitude and meaning of the genetic correlation between the FFW with NSF (rG=0.94**) and TWSF (rG=0.95**) were explained in greater proportion in the genotypic model (rG's) by the direct (PG=-1.66 and PG=1.73) and indirect (1.72 and -1.65) effects. Likewise, the correlation between D1S and FFW (rG=0.68*) was explained via indirect effect (NSF=-1.42 and TWSF=1.47) (Table 3).
In the two types of path analysis (rF's and rG's), a similar trend was observed since the indirect effects via NSF and TWSF stand out but in the opposite direction to explain the degree of significant correlation (p<0.05 and p<0.01) between FFW with FL, FW, FT, V1S, and D1S. A similar situation was observed in the decomposition of the level of association between FFW with NSF and TWSF where the direct and indirect effects between these two traits were important. In general, the results for the two analyses were of opposite signs: positive for NSF values and negative for TWSF in rF's. Whereas, with the use of the rG's, the NSF presented negative values and TWSF positive values (Table 3).
Studies conducted by Lekshmanan & Vahab (2018) pointed out that path analysis with the use of genetic correlations is safer since it does not include environmental effects. This is corroborated in this study, not only with the greater number of significant genetic correlations detected but also because the model for FFW was explained in greater proportion by the genetic correlations since the residual effect (h) for all eight variables investigated was equal to zero (Table 3). This enabled us to deduce that the variation in the level of association of the FFW with the other traits was due more to genetic effects than to the environment. This relationship will allow the breeder to obtain larger fruits and seeds of higher physiological quality, which are essential in the sexual propagation of the species and the generation of seedlings in the nursery.
The importance of the direct and indirect negative effects via NSF and positive effects via TWSF in the path analysis based on the rG's (Table 3) suggests that the TWSF, due to its greater direct, indirect, and positive effect, is the most useful and convenient character in the selection processes of ebony trees, with fruits of greater FFW and therefore greater genetic progress (Pérez-Pelea et al., 2018; Mayavel et al., 2018).
The NSF, although it showed important direct and indirect effects to explain its degree of association with FFW, has a negative value that does not make it a viable trait to obtain genetic gains by selection since it decreases FFW. Similar results have been reported by Agrawal et al. (2018) , who stated that negative coefficients are generally indicative of unfavorable indirect effect on the endogenous variable evaluated. Therefore, whenever possible, any restriction should be made by the NSF or should be discarded in the selection processes.
CONCLUSIONS
Statistical differences were detected for fresh fruit weight; fruit length; fruit thickness; number of seeds per fruit; total weight of seeds per fruit; volume of a seed and density of a seed, which show the existing genetic variability and its potential use in future genetic improvement programs of the species.
Overall, genetic correlations were higher in magnitude and statistical significance than phenotypic correlations. Fresh fruit weight, total weight of seeds per fruit, fruit length, and fruit width were the traits that showed the highest number of significant and positive genetic correlations.
The path analysis based on genetic correlations made it possible to highlight that the total weight of seeds per fruit exerted the most important direct and indirect effects on the degree of association of the fresh fruit weight. Therefore, it is possible to increase the fresh fruit weight directly and indirectly to improve the production and the physiological quality of the seed.


















