Introduction
The genetic modification of plants originated in the early 1980s when new genes of interest were introduced into a plant (i.e., Nicotiana tabacum) using Agrobacterium tumefaciens as a vector (Bevan et al., 1983; Herrera-Estrella et al., 1983; Basso et al., 2020). Since the mid-1990s, the first genetically modified (GM) crops from numerous public and private laboratories have been commercially introduced. In 2019, the area cultivated with GM crops was estimated to be 190.4 million ha in the world, successfully adopted in 29 countries (ISAAA, 2019), with more than 50% of the world's population. The majority of the cultivated area is represented by the most important domesticated species including corn, soybeans, cotton, and canola (Duke & Cerdeira, 2010; Brookes & Barfoot, 2020). According to Brookes (2020), a total of 1.07 million ha have been planted with cotton and corn containing transgenic traits in Colombia since 2003, and farmers benefited from an increase in income of US $301.7 million.
The first patents granted to GM crops have started to expire as these patents are valid for 20 years. These crops are considered off-patent events. These off-patent events compare to generics in the pharmaceutical and agrochemical industry. However, in contrast to the development of the generic industry in the pharmaceutical and agrochemical fields, the lack of harmonization of the current regulatory frameworks of the countries on the sowing, commercial release and use as human and animal food of off-patent crops presents challenges (Rudelsheim et al., 2018).
A GM event has been defined as the insertion of DNA into the plant genome resulting from a single transformation process (Pilacinski et al, 2011; Basso et al., 2020). However, individual GM events may contain one or more transgenes. In other words, a GM event refers to the precise location of an expression cassette in the host genome that encodes a trait of interest (Bell et al, 2018). GM events that confer tolerance to herbicides and/or resistance to insects are the most widely applied engineered traits. These genetic modifications have benefited adopting farmers through higher yields, reduced input costs and less environmental pollution (Carpenter, 2010; Cerdeira et al., 2011; Brookes & Barfoot, 2018).
The timeline for the commercialization of an event is relatively long (the first commercial launch took around 14 years) (Fraley, 2015) and a large investment is needed to comply with all regulatory requirements (McDougall, 2011). In addition, Proprietary Regulatory Property (PRP) holders must obtain regulatory approvals in the countries where they intend to commercially release or export events (Rudelsheim et al, 2018).
Substantial equivalence of a GM crop has been defined as a new product that must be the same as the non-GM crop except for the traits that were enhanced, added, or removed through genetic engineering (OECD, 2002; Parrott et al, 2010; ISAAA, 2018). The concept of substantial equivalence has been used to investigate the compositional safety of transgenic crops (Codex Alimentarius Commission, 2009; Privalle et al, 2013). This approach recognizes that, although no crop or food can be shown to be 100% safe, GM crops can be compared to crops that have a history of safe use (Cheng et al., 2008; Harrigan, Glenn et al, 2010). The most common approach has been to compare the transgenic line with its isogenic version without the transgene, using compositional analysis to identify potential differences in the levels of each of the nutritional components. If no significant differences are found, or are within the expected natural variation, or within ranges previously reported in the literature for each component, the results of this evaluation are presented as a fundamental part of the requirements of the regulatory authorities (OECD, 2003; Harrigan et al., 2010).
Fall armyworm (Spodoptera frugiperda) is one of the most common pests of corn and it has been responsible for large economic losses in Colombia (Gómez et al., 2013; Jaramillo-Barrios et al., 2019). The introduction of insect resistance genes in Colombian maize genotypes is a potential alternative for the control of this harmful pest. For this purpose, the Corn Genetic Improvement Program of the National Cereal and Legume Research Center (CENICEL) of the National Federation of Cereal and Legume Growers (FENALCE), with the support of the research group of Genetic Engineering of Plants of the Universidad Nacional de Colombia (IGP-UN), obtained an off-patent maize hybrid from the TC1507 event. This hybrid was obtained through the backcrossing method using elite Colombian corn lines crossed with Herculex® I corn (GM corn event TC1507), followed by complementary field tests and immunostrip assays (Jiménez et al., 2016). In this way, elite maize lines of the Colombian genotypes with the TC1507 event (cry1fand pat) were obtained. Subsequently, these elite lines with the introgressed event were hybridized to obtain an off-patent maize hybrid with the TC1507 event resulting from the best hybrid combinations of the breeding program (Jiménez et al., 2016). The purpose of the present study was to evaluate the substantial equivalence of this hybrid. The levels of the nutritional components of the proximal analytes in the grain and forage tissues involving the off-patent genotypes (event TC1507) of GM corn plants were compared with the conventional corn genotypes from which they were derived; this hybrid has the same genetic background as off-patent hybrid TC1507 but does not contain the genetic modification.
Materials and methods
Field trial samples
A field trial to obtain grain and forage samples for compositional analysis was carried out. The corn was grown in the CENICEL Paraguaicito experiment station, in the municipality of Buenavista (Quindío), located in the natural subregion of the Colombian coffee zone. This crop had normal pest control and maintenance practices (irrigation, fertilization, herbicide and pesticide applications, etc.), consistent with maize production and applied uniformly to each entire trial area. The planting design consisted of a random block design, with three replicates per block, where six genotypes were distributed: two elite off-patent lines, two conventional elite isolines, one off-patent hybrid (conventional elite line X elite line off-patent), and a conventional hybrid grown locally. From each of the genotypes, three samples of two types of tissues were collected: 1) the samples of the corn grain in its state of maturity (phase R6) and 2) the leaf samples at the same stage. The lines worked were Hybrid Transgenic line (HT), Transgenic line I (T1) and Transgenic line II (T2) of off-patent maize TC1507 event; the non-transgenic control samples lines were Hybrid Conventional (HC), Conventional line I (C1) and Conventional line II (C2).
Compositional analysis
The samples were kept on dry ice until they were transported to the Nutrianalisis laboratory in Bogotá (Colombia) where each of the analytes was quantified. Humidity was determined gravimetrically by placing the samples in a hot-air drying oven at approximately 135°C for 2 h (AOAC method 925.09). Total protein content was determined as total nitrogen using digestion with H2SO4 followed by distillation and titration (Kjeldahl method - AOAC method 979.09). Fat from the corn kernel samples was determined gravimetrically using a Soxhlet extraction with pentane as solvent (AOAC method 922.06). The crude fat of the forage samples was determined gravimetrically by means of an acid hydrolysis procedure (AOAC method 945.02). Crude fiber was estimated using a neutral detergent solution and a thermostable amylase to dissolve easily digestible proteins, lipids, sugars, starches, and pectins. A fibrous residue was obtained which consisted mainly of cell wall components including both nitrogen and indigestible nitrogenous matter (NTC method 5122). Ashes were determined by placing the samples in an electric oven at 550°C until they were incinerated. The residual ash was quantified gravimetrically (AOAC method 923.03). Carbohydrate content was estimated using the calculation previously described by Herman et al. (2004) and Cong et al. (2015).
Statistical analysis
Statistical analysis of nutrient composition data was performed using R software (R Core Team, 2021). To perform the statistical analysis, four functions were written. The first function, which was the basis for the other functions, allowed the generation of a database for graphic analysis and statistical tests of differences between groups. The second function performed an exploratory data analysis and obtained a box-and-whisker plot resulting from each analyte per genotype. The third function established the confidence intervals of each analyte according to the Composition Database of the International Institute of Life Sciences (Ridley et al, 2004; AFSI-CCDB, 2020). Finally, the fourth function performed the difference tests between the genotypes according to each analyte analyzed. The database includes data from the evaluation of conventional crop samples using validated analytical methods, providing a robust collection of high-quality non-gene compositional data for various crops (Suit et al., 2016). The statistical tests performed included the Shapiro-Wilk test for normality, the Levene test for homoscedasticity and the Kruskal-Wallis test for differences between groups and their respective Wilcoxson post hoc test (Bonferroni test). Two separate statistical analyzes of the compositional data were performed. The quantification of the analytes made it possible to evaluate the data of the replicas of each of the corn grain and forage analytes using the Kruskal-Wallis test to identify statistically significant differences (α = 0.05) between the genotypes and their respective Wilcoxon post hoc test. This evaluation allowed corrections on the level of significance to avoid increasing the type 1 error and to identify the significant differences between each of the genotypes (α=0.05).
Results and discussion
Grain analytes
The results of the proximate analysis from the grain samples collected during the field trial did not show statistically significant differences between the transgenic genotypes [Hybrid Transgenic (HT), Transgenic line I (T1) and Transgenic line II (T2)] of off-patent maize TC1507 event and the non-transgenic control samples [Hybrid Conventional (HC), Conventional line I (C1) and Conventional line II (C2)] (Fig. 1). For instance, the Kruskal-Wallis test showed differences for grain components such as moisture (P-value=0.0323), protein (P-value=0.0093), fat (P-value=0.0237), and carbohydrates (P-value=0.0085) in at least two of the genotypes in comparison. However, after performing a post-hoc test, the Mann-Whitney test (Wil-coxon rank sum test) did not show significant differences between the genotypes for all analytes that initially seemed to differ. The Wilcoxon rank sum test corrects the level of significance to avoid increasing the type I error and shows in which groups there are differences.
Figure 1 shows that, for each of the analytes corresponding to each genotype, there are no differences with the maximum and minimum ranges reported by the literature (Ridley et al, 2004; AFSI-CCDB, 2020).

FIGURE 1 Variability of percentage values for proximate analytes from corn grains. A) Moisture; B) Protein; C) Total fat; D) Crude fiber; E) Ash; and F) Carbohydrates. [Hybrid Transgenic (HT), line Transgenic line I (T1) and line Transgenic line II (T2)] of off-patent maize (TC1507 event) and the non-transgenic control samples [Hybrid Conventional (HC), Conventional line I (C1) and Conventional line II (C2)]. Values are expressed as medians (±IQR, n=3) from three independent experiments. * indicated the P-value < 0.05 for Kruskal-Wallis test.
For each of the different analytes studied (Fig. 1 A, D), the variation for moisture and fiber is very similar for each of the compared genotypes, although there are slight differences for other analytes (Fig. IB, C, E, F). For total fat, the conventional hybrid tends to have higher levels than the transgenic hybrid. The comparison of the lines shows unexpected data: the transgenic line presents a variation similar to that of the comparable non-transgenic line. For protein, the transgenic hybrid presented the lowest values together with the conventional hybrid, while the CI had the highest values in contrast to its transgenic isoline and the T2 and C2. For ash, only the CI presented a great variation unlike the other genotypes analyzed that maintained a similar variation. For carbohydrates, the transgenic hybrid had higher levels, while the CI showed lower values. The remaining genotypes maintained similar variations. All these apparent differences are within natural variations. All the values for analytes examined were found within the values reported in the literature (Watson, 1982; Lundry et al, 2013; Cong et al, 2015; AFSI, 2020) (Tab. 1).
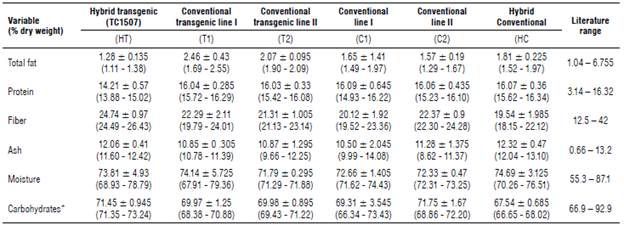
TABLE 1 Medians, minimum and maximum values reported from the proximate grain analysis of off-patent maize hybrid TC1507 (transgenic), off-patent maize lines TC1507 I and II (transgenic), their respective conventional isolines I and II (non-transgenic) and a commercial maize hybrid (non-transgenic) from the samples collected from the field trials in Colombia.

* Watson (1982), Lundry et al. (2013), Cong et al. (2015), Anderson et al. (2019), AFSI (2020).
Forage
The results of the proximate analysis for the forage samples collected during the field trial did not show statistically significant differences between the transgenic genotypes (HT, line T1, and line T2) of off-patent maize event TC1507 and the non-transgenic control samples (HC, line C1, and line C2). The Kruskal-Wallis test showed differences only for the fat component (P-value=0.03), finding that at least two of the genotypes in comparison exhibited differences. However, after performing a post-hoc test, the Mann-Whitney test (Wilcoxon rank sum test) showed no significant differences between the genotypes (Tab. 2).
TABLE 2 Medians, minimum and maximum values reported from forage proximate analysis of off-patent maize hybrid 1507 (transgenic), off-patent maize lines 1507 I and II (transgenic), their respective conventional isolines I and II (non-transgenic) and a commercial maize hybrid (non-transge-nic) from samples collected in field trials in Colombia.
* Watson (1982), Lundry et al. (2013), Cong et al. (2015), Anderson et al. (2019), AFSI (2020).
There were no cases in which the analyzed analytes showed differences in their maximum and minimum ranges with those reported in the literature (Fig. 2). For moisture, protein, fiber, fat, ash, and carbohydrates, the observed variations were very similar between each of the compared genotypes, although at first glance there are slight differences. For moisture, only the HT presented lower values than the other genotypes, which presented values slightly above the maximum limit reported in the literature (Fig. 2A). For fat, the HT presented lower values and T1 higher values compared to the other genotypes (Fig. 2C). For crude fiber, only the hybrids showed slight differences, while the conventional and the transgenic lines showed lower and higher values, respectively (Fig. 2D). For moisture, differences were not observed, although greater variations were observed for both the HT and the Tl. For ash, the HC showed slightly higher values compared to the other genotypes (Fig. 2A and E, respectively). For carbohydrates, the conventional hybrid showed slightly lower values than the other genotypes (Fig 2F). The conventional hybrid had a carbohydrate value lower than that reported in the literature, while its other values (highlighting the median) were found within the ranges reported. All these apparent differences were found within natural variation. All the values for the analytes examined were found within the values reported in the literature (Watson, 1982; OECD, 2002; Lundry et al., 2013; Cong et al., 2015; Anderson et al, 2019; AFSI, 2020) (Tab. 2).

FIGURE 2 Variability of percentage values for proximate analytes from corn forage. The methods used for estimating the percentage values for each genotype are described in the Material and methods section. A) Moisture; B) Protein; C) Total fat; D) Crude fiber; E) Ash; and F) Carbohydrates. [Hybrid Transgenic (HT), line Transgenic line I (T1) and line Transgenic line II (T2)] of off-patent maize (TC1507) event and the non-transgenic control samples [Hybrid Conventional (HC), Conventional line I (C1) and Conventional line II (C2)]. Values are expressed as medians (± SD, n=IQR, n=3) from three independent experiments. * indicated the P-value < 0.05 for Kruskal-Wallis -test.
Analysis strategy of the substantial equivalence of off-patent corn
The developers of different transgenic events, both simple and stacked, have obtained authorizations in Colombia for human and animal consumption using data portability, since the trials in most cases were not carried out in Colombia but in northern agroecosystems (ICA, 2011, 2013; MPSP, 2012a, 2012b, 2012c, 2014; INVIMA, 2018). These developers have performed substantial equivalency assessments based on the compositional analysis of proximate analytes.
Based on the principle of equality, the strategy for off-patent crops with transgenic events that have been previously authorized in Colombia could be incorporated into conventional breeding since it is in the public domain (its use does not violate the rights of third parties). In addition, the analysis of freedom of operation (Jiménez et al., 2016; Rojas et al., 2017) makes it possible to carry out field trials with a minimal design, where a limited number of proximate analytes equal to the number examined by developers of the specific event that allowed to support the substantial equivalence of stacked events that contain the same event.
The case of stacked events (stacked traits or crop stacking) originating from conventional plant breeding, where two or more parents with a single transgenic event are crossed to produce a progeny containing two or more transgenic events that provide a useful grouping of traits would be the closest issue to off-patent events. The discussion that has arisen today could help resolve the regulatory gap and allow the authorization for human and animal consumption of these derived crops, where substantial equivalence and other safety assessments play a fundamental role (Kok et al, 2014). From a regulatory point of view, stacked trait and off-patent crops differ from single trait crops, since they contain events that are not new to regulatory authorities considering that they have been fully evaluated in the single events that make them up. This distinction is crucial since the concern for the safety of transgenic crops is centered around the safety of the introduced trait and the possibility of unintended adverse effects from its introduction (Acosta & Chaparro, 2008; Codex Alimentarius, 2009; Herman & Price, 2013).
Safety assessment approaches for stacked trait crops vary globally. There are regulatory bodies that address a crop of stacked traits as if it were a crop with a new event that requires a de novo assessment, despite the existence of evidence and safety evaluations demonstrating the safety and substantial equivalence for each one of the individual transgenic events that make up the crop with stacked events. This approach is based on the concern about the possible interactions between events that could give rise to characteristics of the plant different from the expected sum of the stacked events. Steiner et al. (2013) extensively analyzed the overall potential for event interactions in stacked event crops. The authors highlighted that since the functional characteristics of the introduced expression cassettes are known, it is possible to develop hypotheses as to whether a specific combination of traits would interact to affect plant metabolism in a novel way. It was also pointed out that if a hypothetical interaction posed any risk, for example as in the case of stacking of tolerance traits to glyphosate and dicamba through conventional plant breeding, the molecular mechanisms of the enzymes that mediate these stacked tolerances would exclude any plausible interactions. In fact, the substantial equivalence of a crop with stacked traits that confers tolerance to glyphosate and dicamba has been empirically demonstrated (Taylor et al., 2017). Specifically, for corn containing traits of resistance to insects and tolerance to glufosinate-ammonium, there is no plausible hypothesis that their combined presence affects the metabolism of the plant and leads to compositional differences. Likewise, it is even less plausible that undesirable interactions occur in an off-patent obtained from the crossing between a transgenic hybrid and a conventional maize line.
In the global landscape of regulations on genetically engineered crops, there are currently two general approaches to assess food and feed safety for stacked event crops that could be applied for off-patent crops. In the first approach, no additional data or evaluations are required for a stacked trait crop generated by conventional plant breeding whose constituent events have previously been evaluated and approved. Regulatory agencies may require written notification for the stacked event product to be marketed. Examples of agencies that follow this approach include the FDA and APHIS-USDA of United States, the Canadian Food Inspection Agency, the Canadian Ministry of Health, the Australian and New Zealand Food Standards Agency, and the Food Safety Commission and the Ministry of Agriculture, Forestry and Fisheries of Japan (Pilacinski et al., 2011; Steiner et al., 2013; FSCJ, 2016; Goodwin et al., 2021). In the second approach, a safety assessment is required for any stacked trait crop, regardless of whether its constituent events were previously assessed and approved. Some examples of regulatory agencies that follow this approach are the European EFSA, the Ministry of Food and Pharmaceutical Safety of the Republic of Korea, the Food and Drug Administration of Taiwan, and the Federal Commission for the Protection against Sanitary Risks of Mexico (Pilacinski et al., 2011; Steiner et al., 2013; FSCJ, 2016; Goodwin et al., 2021).
In rational and scientifically sustainable risk assessment (Pilacinski et al., 2011; Steiner et al., 2013; FSCJ, 2016; Goodwin et al., 2021), it is important to follow the case of stacked trait crops that could contribute to solving the regulatory challenge of off-patent crops. In this context, the formulation and evaluation of a risk hypothesis for a crop in question is central in determining whether further evaluation of the safety of a product with stacked traits is needed.
Today there is a growing body of empirical data in reports and/or in scientific articles evidencing the safety of genetically modified crops of relevant single events as well as the safe historical use of conventional plant breeding. Therefore, there is valid scientific justification to eliminate or make more flexible the mandatory requirements aimed at presenting and evaluating characterization, safety, and substantial equivalence data for each new crop of stacked events (Goodwin et al., 2021) and, therefore, for off-patent crops. Pragmatically, as proposed by Bell et al. (2018) and Goodwin et al. (2021), a gradual approach could start with flexibility and end with the elimination of the requirements for safety assessments for the crops of stacked events (and therefore off-patent crops). Along these lines, the first step could be to refine the approach towards a single specific analysis of stacked events. This approach would provide an empirical bridge to the previous evaluations performed for the approved transgenic events that make up the stacked events. In this way, a simplified evaluation of the stacked event or of the off-patent crop would focus on the composition, evaluating the substantial equivalence from the proximate analytes of the grain (and from the forage, in the case of resistance to insects) in the context of known natural variability. Such an assessment could be used to complement previously reached safety conclusions for constitutive individual trait products (Bell et al., 2018; Goodwin et al., 2021).
The comparative analysis of corn during field trials carried out in Colombia showed no significant differences between off-patent hybrids of lines 1507 I and II obtained by conventional crossing with their non-genetically modified counterparts. Small exceptional variations were not indicative of a general pattern of changes derived from crossing with a genetically modified hybrid; these are explained by the well-known natural variation (genotype-environment interaction). These variations in composition arise due to a wide range of factors specific to the biology of the species and, particularly, to the growing conditions (location, irrigation, type of soil, etc.) that influence the growth and biochemistry of plants. In the present study, no significant biological differences were found between off-patent TC1507 event corn and non-transgenic conventional corn. Therefore, the off-patent hybrid is substantially equivalent from a compositional point of view to its conventional counterpart, except for the genetically engineered characteristics. The composition of the off-patent TC1507 corn hybrid was found within the normal ranges of variation observed in non-genetically modified varieties with recent biotechnological tools.














