Introduction
Tuberculosis, which is caused by Mycobacterium tuberculosis, is one of the deadliest infectious diseases in the world and re presents a public health problem1. In recent years, the number of strains of M. tuberculosis resistant to first-line drug, such as rifampicin, has been increasing. Rifampicin plays an important role in the first line of tuberculosis treatment due to its powerful bactericidal effect. This drug inhibits the synthesis of messen ger RNA by binding to RNA polymerase. Rifampicin resistance is caused by single nucleotide polymorphisms (SNPs) on rpoB gene, which encodes the beta subunit of RNA polymerase2.
On the other hand, ponA1 gene encodes the penicillin bin ding protein PonA1, which participates in peptidoglycan biosynthesis and regulates the enzymatic activity of other proteins involved in bacterial morphology and growth3,4. Some SNPs (T34D and Q365H) have been reported to alter the tolerance of M. tuberculosis to rifampicin and increase the minimum inhibitory concentration (MIC) almost twice5,6. Likewise, a recent study found that some SNPs (P631S and A516T) are associated with rifampicin resistance and su ggested these SNPs could alter the rifampicin tolerance of M. tuberculosis. Nevertheless, a causal association between rifampicin resistance and the presence of these SNPs could not be established7. For these reasons, this study included the ponA1 gene in the analysis since we consider that it plays a role in rifampicin resistance, and could be associated with some spoligotypes defined lineages.
At the end of the last century, a new method was developed for the genotyping of M. tuberculosis strains, which was called spacer oligonucleotide typing or spoligotyping, based on the presence or absence of certain spacer sequences in the direct repeat region (DR). The DR region is a DNA segment made up of a 36 bp repeat sequence and several 31 to 41 bp non-repeating DNA segments called spacer sequences. 43 spacer sequences were identified in M. tuberculosis H37Rv and made it possible to group the strains of this bacteria into certain lineages8. Several studies have reported an association bet ween the presence of certain lineages identified by spoligo typing and resistance to certain drugs9-11; for example, most of the Beijing lineage M. tuberculosis strains that have been studied presented drug resistance and this frequency was higher compared to strains of other lineages. Therefore, a probable causal association between the presence of certain lineages and drug resistance has been suggested; however, this probable causal association could not be confirmed12.
Our study hypothesizes that some lineages would be asso ciated with the presence of SNPs in genes associated with drug resistance such as rpoB gene and genes involved in some drug tolerance mechanism such as ponA1 gene. In this sense, this study analyzed two secondary database of 484 M. tuberculosis genomes in order to identify an association between the presence of SNPs on rpoB and ponA1 genes and the presence of certain spoligotypes defined lineages.
Materials and methods
Our study performed a statistical analysis of a secondary da tabase, which comes from two previous studies13,14 of 484 ge nomes obtained from strains of M. tuberculosis, which were isolated from patients with active tuberculosis in the cities of Lima and Callao. The genomic DNA extracted from M. tuber culosis was sequenced using Illumina HiSeq2000. High quali ty sequencing reads were assembled taking into account the genome of the reference strain M. tuberculosis H37Rv using NextGENe software. The alignment of the sequences with this reference genome made it possible to identify SNPs, inser tions and deletions using NextGENe software. These studies identified the lineage (clade and SIT code) of each strain13,14.
The rpoB and ponA1 genes were classified considering each lineage separately as an exposure variable and the presence or absence of at least one SNP on rpoB and ponA1 genes, both genes were studied separately, as outcome variables. This analysis was performed to determine the association between lineages defined by spoligotypes and the presence of at least one SNP on rpoB and ponA1 genes or the absence of SNP in these genes (Table 1).
Where “X” can be rpoB gene or ponA1 gene. “Y” represents any of the lineages included in this study.
Our study also carried out an analysis where it considered rifampicin resistance as an outcome variable (table 2). Fur thermore, we included in our study SNPs with a frequency greater than 5% of the ponA1 gene, for which SNPs were considered separately as outcome variables.
“Y” represents any of the lineages included in this study.
The odds ratio (OR) was then calculated for each lineage in cluded in this study. A significance level of 95% was consi dered and the statistical calculations were carried out using programming language R.
Results
Approximately 63.64% of the genomes analyzed in this study presented at least one SNP in ponA1 gene and 72.52% pre sented at least one SNP in rpoB gene. Therefore, it could be inferred that the SNPs of these genes are relatively frequent in M. tuberculosis strains isolated from patients with active tuber culosis in the cities of Lima and Callao. Furthermore, 71.69% of these genomes come from rifampicin-resistant strains.
Our study found that the analyzed genomes was grouped into 4 lineages (LAM, Haarlem, T and Beijing). If we consi der the classifications described in the methodology, we can observe that the LAM lineage contained the highest number of genomes in the three proposed classifications. However, the Haarlem lineage was presented in approximately 29.9% of genomes with at least one SNP on ponA1 gene (figure 1). Also 16.2% and 15.9% of genomes with at least on SNP on rpoB gene (figure 2) and genomes derived from rifampicin-resis tant strains (figure 3), respectively; were found in the Haarlem lineage. Only 4.2% of genomes of the Haarlem lineage did not present at least one SNP on ponA1 gene. Hence this fact could be suggesting that there is an association between the presence of SNPs on ponA1 gene and the Haarlem lineage.
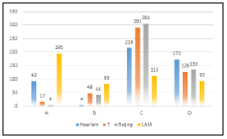
Figure 1 Frequency of spoligotypes defined lineages considering the presence or absence of at least one SNP on ponA1 gene.
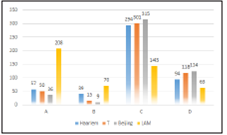
Figure 2 Frequency of spoligotypes defined lineages considering the presence or absence of at least one SNP on rpoB gene.

Figure 3 Frequency of spoligotypes defined lineages considering the considering resistance or susceptibility to rifampicin.
This study found a statistical association between the presen ce of at least one SNP on ponA1 gene and the Haarlem and LAM lineages (ρ-value<0.0001). Likewise, the absence of SNPs on ponA1 gene is associated with the T and Beijing lineages (ρ-value<0.001) (table 3). Furthermore, the rifampicin resistance is associated with the LAM lineage (ρ-value<0.01) (table 3).
Table 3 Statistical analysis of rifampicin resistance, rpoB and ponA1 genes.

*Statistical significance. CI: Confidence Interval.
Our study found an association between the presence of P631S SNP and the Haarlem and LAM lineages (ρ-value <0.0001). Likewise, SNP A516T SNP presented an association with the LAM lineage (ρ-value <0.001) (table 4). Other SNPs of the ponA1 gene were not included in the analysis since their frequency was less than 5% of the total SNPs.
Discussion
Since the advent of spoligotyping, it has been possible to analyze the spacer sequences of the DR region of M. tubercu losis strains, all over the world. Which has provided conside rable information on the distribution of the lineages defined by spoligotypes12. In this sense, various studies have found a possible association between certain lineages and resistance to first-line drugs9,11. Which has opened a debate on whether these associations are causal or results of chance12. Our study evaluated whether the presence of at least one SNPs on rpoB and ponA1 genes or rifampicin resistance are associated with some spoligotype defined lineages.
More than 96% of rifampicin resistant strains have been reported to have SNPs in an 81bp region of rpoB gene15. Likewise, studies conducted in China and Russia have found a probable association between rifampicin resistance and the Beijing lineage16,17. An association between this lineage and multidrug-resistant tuberculosis (MDR-TB) has also been found in India and Vietnam9,18. Other lineages have also been associated with rifampicin resistance12; for example, a stu dy in Brazil reported the association between the LAM and T lineages and resistance to rifampicin and isoniazid19. Our study found that LAM lineage is associated with rifampicin resistance and the absence of SNPs on rpoB gene is associa ted with the Haarlem lineage. We did not find a statistically significant association between the presence of at least one SNP in rpoB gene and the lineages included in the study. This could be due to the fact that there is no causal relationship between rifampicin resistance and the most frequent linea ges reported in Peru. A study carried out in Peru did not find an association between resistance to drugs, including rifam pin, and the lineages found in this country20. However, we found an association between rifampicin resistance and LAM lineage. This could be due to the fact that not all SNPs in the rpoB gene cause resistance to rifampin and there is almost 5% of cases of resistance to this drug that are not associa ted with the rpoB gene. Other studies, conducted in Mexico, Uganda and Italy also found no association between drug resistance and spoligotype defined lineages 21,22,23.
On the other hand, in vitro studies have reported that some SNPs (T34D and Q365H) on ponA1 gene increase the MIC of rifampicin to almost double, compared to strains of wild-type M. tuberculosis5,6. Likewise, a study analyzed 914 genomes of M. tuberculosis from Peruvian strains and found that SNPs P631S and A516T are associated with resistance to this drug; this study found that SNP A516T is more common in rifampicin-resistant strains7. Our study found that the presence of at least one SNP on ponA1 gene is associated with the LAM and Haarlem linea ges and the absence of SNPs on this gene is associated with the Beijing and T lineages. Furthermore, we found that the most common SNP in ponA1 gene (P631S) is associated with LAM and Haarlem lineages. The A516T SNP also is associated with LAM lineage. Our study suggests that this could be due to the fact that the strains of this lineage developed in a medium that allowed the selection of SNPs on ponA1 gene, especially SNPs that alter bacterial morphology and growth. It has been sugges ted that the SNP P631S could generate these alterations; likewi se, it was the most frequent SNP found in Peru7.
On the other hand, associating the absence of a SNP to a group, as this study did, might not be appropriate since we could be dealing with a conserved gene or with low evolutio nary pressure. However, the genes evaluated present a high frequency of SNPs both in our study and in previous studies.
Most of the genomes included in our study come from MDR-TB strains; hence, our results could be due to resistance to other drugs or related to genes associated with other drugs. However, we present the first evidence that suggests an as sociation between the presence of at least one SNP on ponA1 gene and the LAM and Haarlem lineages.