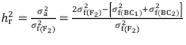INTRODUCTION
Watermelon [Citrullus lanatus (Thunb.) Matsum and Nakai] is one of the highest yielding fruit crops in Brazil, and according to the 2016 world ranking, the country was ranked as the 4th largest watermelon producer, with 2,090,432 t of fruit, second only to China (79,043,138 t), Turkey (3,928,892 t) and Iran (3,813,850 t) (Fao, 2018). The development of new genotypes with commercial attributes different from those already on the market, such as the size of the seeds, can make the production of Brazilian watermelon more expressive. Moreover, the consumer market has increasingly been demanding nutritious and healthy fruits (Farias et al., 2014).
For fresh consumption, the smaller the seed size, the easier it becomes to consume the fruit, while for the production of fruit and sowing in the field, the larger the seed size is, the greater the production and the easier the planting. Thus, understanding the genetic control of traits is important for the development of genotypes aimed at having the most diverse characteristics (Li et al., 2018). However, it is worth mentioning that information about the genetic behavior of watermelon characteristics is scarce. Among these reports, those studying the size of the seeds, besides being rare, are quite old, which characterizes it as being one of the main obstacles for breeders.
In previous studies, Poole et al. (1941), cited by Li et al. (2018), described that the genetic control of seed size was controlled by two genes, 's' and 'l', with 's' being epistatic to 'l'. A few years later, the 'Ti' genes for tiny seeds and 'ti' genes for tomato seeds were described (Tanaka et al., 1995; Li et al., 2018). According to Yong-Jae et al. (2009), watermelon seed size can be classified as giant, large, medium, small, micro and tiny. In addition to seed size, inheritance studies of other characteristics in watermelon have been reported, including fruit yield (Kumar and Wehner, 2013), seed mass (Adjouman et al., 2016), resistance to the PRSV-W virus (Alves et al., 2014), and egusi seed (Gusmini et al., 2004). This suggests that before performing a characteristic improvement, it is necessary to understand the genetic control mechanisms within the populations of interest.
Thus, the objective of the present work was to study the inheritance of seed size characteristics in watermelon populations through crossing of contrasting genotypes.
MATERIALS AND METHODS
Obtaining the populations
The experiment was conducted from 2017 to 2018 at the Bebedouro Experimental Station at Embrapa Semiárido in Petrolina-PE (Brazil). Two watermelon lines, with contrasting seed size, were used line 31715.001 (“P1”) (small seed) and lineage 31717.005 (“P2”) (large seed), to obtain F1, F2, BC1 and BC2 populations.
As such, crosses were carried out via controlled hand-pollination (CHP) according to the methods used by Nascimento et al. (2018). Regarding the CHP, in the flowering stage, male and female flowers of the lineages and the F1 hybrids were isolated at preanthesis using aluminum sachet bags. On the following day, the open male flowers were detached from the plants, their petals were removed to expose their anthers, and then pollen grains were gently deposited on the stigmas of the female flowers. After this procedure, the pollinated flowers were identified with cardboard tags containing information on the parental crossings, plant type, and CHP date. Afterward, the flowers were isolated again for a period of 72 h. Between 30 and 35 d after pollination, the fruits were harvested, and their seeds were extracted, washed and left to dry in the shade.
To obtain the F1 population, between January and April 2017, seeds were sown in polystyrene trays containing proper commercial substrate and then kept in a greenhouse for 12 d. After that period, the seedlings were transplanted to the field.
In the following generation, five seeds from the parents (P1 and P2) and from the F1 population were sown during the months of May to August 2017, and using CHP, the F1 plants were self-fertilized and simultaneously crossed with the parents, P1 and P2, which produced the seeds of F2, BC1 and BC2 populations. All populations obtained were then evaluated in the field, from May to August 2018, under open pollination.
Fertilization was carried out according to a soil analysis and as recommended by Mendes et al. (2010), with 30 kg ha-1 of N, 80 kg ha-1 of P2O5, and 30 kg ha-1 of K2O, plus 15 kg ha-1 of ZnSO4 and 10 kg ha-1 of CuSO4. Fertilizers were top-dressed with irrigation water containing 50 kg ha-1 of N in the form of Ca (NO3)2 and 40 kg ha-1 of K2O using KCl, between 50 to 60 d after sowing.
The water was applied by drip irrigation at a daily depth that varied according to the plants’ needs depending on climatic conditions, monitored by a weather station located near the experimental area. The crop coefficients (Kc values) used to calculate crop evapotranspiration (ETc) were those obtained by Freitas and Bezerra (2004) in Canindé, CE, Brazil, for Crimson Sweet watermelon, whose corresponding values were 0.46 to 0.70 at the vegetative stage, 0.89 to 1.22 at the flowering stage, and 1.14 to 0.74 at the fruiting stage.
Phytosanitary treatments were carried out with agrochemicals registered by the Brazilian Ministry of Agriculture, Livestock and Supply, and weeds were removed manually.
Phenotypes of seed size in watermelon populations
During the months of April to August 2018, seeds from all the populations were germinated in expanded polystyrene trays containing commercial substrate for vegetables. At 12 d after sowing, the seedlings were transplanted into the field at the Bebedouro Experimental Station in accordance with a completely randomized design, with each plant considered an experimental unit and with a varied number for each population, in soil previously prepared with gray plastic mulching sheets, with 1 m spacing between plants and 2.5 m between rows. After transplanting, the seedlings were covered with an agrotextile blanket until the flowering period, at which point it was removed for pollination. The seeds of each population came from a single fruit.
At 70 d after planting, fruits were harvested. The seeds were subsequently extracted, washed and left to dry in the shade. Eight, 10, 7, 271, 95 and 94 plants from the P1, P2, F1, F2, BC1 and BC2 populations, respectively, were evaluated.
To investigate seed size inheritance, seed length was measured, as this characteristic is highly correlated with width and weight (Hawkins and Dane, 2001). The lengths of 10 seeds were measured with a digital caliper (Stainless Hardened: JL-YB5474) following the method used by Nascimento et al. (2018), with seeds randomly chosen from one fruit of each plant.
The mean and variance [P1
 , P2
, P2
 , F1
, F1
 ; F2
; F2
 , BC1
, BC1
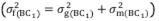 and BC2
and BC2
 ] of the populations were then estimated.
] of the populations were then estimated.
The mean degree of dominance (MDD) of the characteristic was estimated based on the variances and in accordance with the model MDD =  . Broad-sense (
. Broad-sense (
 ) (1) and narrow-sense (
) (1) and narrow-sense (
 ) (2) heritability were estimated as follows:
) (2) heritability were estimated as follows:
Additionally, the values of k1 (based on variance) and k2 (based on the mean) were estimated, where k = 0 indicates the absence of dominance (k=0=additive), 0 ≥ k ≤ 1 indicates incomplete dominance, k>1 indicates over dominance, and k=1 indicates complete dominance.
Additionally, the minimum number of genes (MNG) controlling the characteristic was estimated by the formula n =  . In terms of computer resources, the “segregating and non-segregating generations” procedure of the GENES software program was used (Cruz, 2016).
. In terms of computer resources, the “segregating and non-segregating generations” procedure of the GENES software program was used (Cruz, 2016).
Afterward, the characteristic mean was subjected to the chi-square (X 2) test at a 0.05 significance level for verification of the mendelian pattern within the F2 population according to the following model (3):
where X² is the chi-square value calculated and Obs i and Esp i are the observed and expected frequencies of the i-th phenotypic class (i = 1, 2, ..., n), respectively (Schuster and Cruz, 2013).
RESULTS AND DISCUSSION
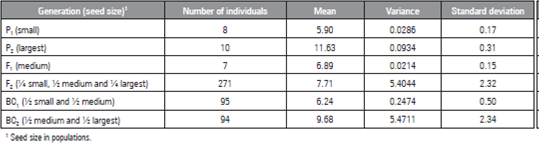
The mean, variance and standard deviation of the seed length of the parents of the segregating populations (Tab. 1) were different, which indicated the existence of genetic variability between them.
Table 1 Number of plants, means, variance and standard deviations of seed size, as evaluated for six watermelon populations.
The results are in line with the assumption of Cruz et al. (2014), who stated that better precision in genetic analyses requires parents whose studied characteristics contrast. According to those authors, genetic parameter estimates allow the identification of the nature of the action of the genes involved in characteristic control. Thus, estimates can be used as an advantageous auxiliary tool in breeding program management.
The variance expressed by the parents and the hybrids showed low values, while the F2 and RC2 populations presented high values. Similar results were reported by Souza et al. (2006), who, when studying seed size inheritance in watermelon generations, observed high values of variance in the F2 and BC2 populations. The high values in both studies possibly occurred because gene segregation within these populations is higher than in the other populations.
Although watermelon is an allogamous species, the low values of variance in the P1, P2 and F1 populations of the present study are due to environmental factors, since the parents used to develop the segregating populations were generated by seven cycles of self-fertilization. Thus, each of the plants of these generations is considered to correspond to the same genotype. However, expecting higher values of the variance of F2 populations and backcross generations due to the interaction of genetic and environmental factors influences the phenotypic expression of traits (Cruz et al., 2014).
The F2 population presented the fourth lowest mean. According to Zewdie and Bosland (2000), depending on the interaction of a trait, the mean for this population may increase or decrease, indicating that parents are contributing alleles to increase or decrease the trait under study. Based on this information, the mean value of the F2 population may have occurred because most seeds were classified as medium and short, which together represent approximately 75% of the total amount.
According to the genetic parameter estimates for the F2 population (Tab. 2), in comparison with the environmental variance, the genetic variance had the greatest influence. Nascimento (2017) found similar results for watermelon seed size, with a higher genetic variance than environmental variance. These results show that the environment has little influence on the phenotypic expression of the evaluated characteristic and that the phenotype observed was mostly the result of genetic influence.
Table 2 Genetic parameters related to seed size in a watermelon F2 population obtained by crossing contrasting varieties.
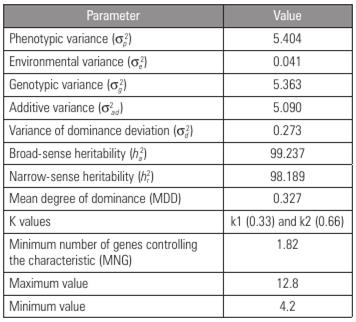
The estimates of the phenotypic, genotypic, additive and environmental variances showed that the contribution of the additive-effect genes to the dominance genes in terms of seed size was significant (Tab. 2), indicating additive allelic interaction. Corroborating the results of the present study, Tanaka et al. (1995), when examining the inheritance of fruit shape and seed size in two contrasting watermelon populations, also observed that seed size was governed predominantly by additive allelic interaction, with the small-seed phenotype being an easily improved characteristic.
However, Adjoumani et al. (2016), when studying the genetics of seed traits resulting from the intraspecific crossbreeding of distinct watermelon varieties, observed that the effects of genes for the characteristic in question showed dominant allelic interactions. The differing results reported by the above authors can be attributed to the genetic differences of the populations evaluated in each study.
It is worth emphasizing that the predominance of additive allelic interaction facilitates the selection stage, since superior individuals also show superior descent (Cruz et al., 2014). Regarding dominant allelic interactions, breeding programs can benefit from developing hybrids.
Within the F2 population, one superior individual was observed for the smallest seed size (4.2 mm) (Tab. 2). This individual can be further investigated and give rise to a small-seed open-pollinated cultivar and can also produce a small-seed-size variety, which can be used for hybrid combinations that may result in reduced-seed-size genotypes. It is worth noting that small seed size is dominant over large seed size, and medium size is dominant over small and large sizes (Guner and Wehner, 2004).
The use of transgressive genotypes to develop hybrids with reduced seed size can be very promising, as producers may be able to acquire hybrid seeds of relatively large size, making planting easier, whereas the fruits of these hybrids will have relatively small seeds, thus making watermelon consumption more agreeable and providing a market advantage over the other genotypes.
In the present study, individuals from the F2 population presented the following approximate phenotypic percentages of seed size: small (25%), medium (50%) and large (25%). Previous inheritance studies on watermelon seed traits such as weight showed that monogenic segregation at a 3:1 ratio did not occur as expected but was close enough to suggest that the smallest seed mass was monogenic and dominant over the largest seed mass (Weetman, 1937).
Similar results were observed in the present study for seed size, where the smallest size, although not in accordance with the expected 3:1 ratio, was quite close to a ratio of 1:2:1, thus suggesting that the characteristic is controlled by two genes.
In addition, the seed size characteristic showed high broad-sense and narrow-sense heritability (Tab. 2). Similar results were observed by Amburani (2018), who, when studying the heritability of morphoagronomic traits in 30 watermelon genotypes, obtained heritability estimates of more than 80% for seven of the 13 traits evaluated, including 100-seed weight and number of seeds per fruit. According to Cruz et al. (2014), narrow-sense heritability represents the variance of the additive type fixed in the populations as a result of the advancement of generations. Based on this information, the correlation between phenotypic values and genetic values is considered high for the characteristic in question, and it is possible to select individuals with relatively small seed sizes based on their phenotype.
To infer the mean degree of dominance and determine the best strategy to improve various characteristics, the values of k1 (0.33) and k2 (0.66) (Tab. 2) were calculated and showed mutual agreement, indicating the existence of incomplete dominance for seed size in the studied populations. Adjoumani et al. (2016), when studying the genetics of seed traits resulting from the intraspecific crossing of distinct watermelon varieties, obtained the same results. In both studies, these results reinforce the efficiency of genotype selection of individuals of interest based on the phenotype.
The minimum value for the number of genes involved in the control of the studied characteristic was 1.82 (Tab. 2). This information is important for estimating the type of inheritance that controls a given characteristic (Lobo et al., 2005). In the present study, seed size is controlled by two genes, indicating that simpler breeding methods, such as mass selection, can be used to obtain small-seed-size cultivars.
When we performed the chi-square test on the populations (Tab. 3), there were no significant differences between populations for observed and expected frequencies associated with phenotypic segregation of the BC1 population, in which all individuals should present the same seed size. However, it was observed that the seeds of the individuals were divided into small and medium sizes, confirming the hypothesis by which two genes control the characteristic under incomplete dominance.
Table 3 Chi-square test results of the watermelon populations to investigate the segregation of two loci based on the 1:2:1 ratio.
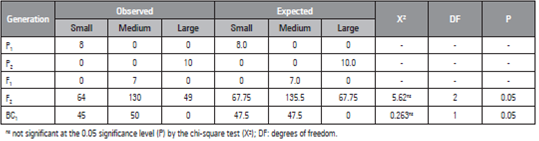
Similarly, Poole et al. (1941), cited by Prothro et al. (2012) studied seed length inheritance by evaluating crosses between plants that produce small and large, small and medium, and medium and large seeds and proposed that this characteristic is controlled by two genes because medium-size seeds were dominant over small- and large-size seeds. Those authors proposed that the recessive l and s genes afforded large- and small-seed phenotypes, respectively. In addition, among the evaluated genotypes, the s gene was epistatic for l, and LL SS was assigned to medium-size seeds; ll SS, large-size seeds; and LL ss or ll ss, small-size seeds.
On the other hand, Tanaka et al. (1995) proposed that, in a Sweet Princess watermelon population, tiny seed size was monogenic, controlled by the Ti gene and dominant over medium seed size; thus, the tiny seed size characteristic was possibly governed by simple inheritance.
The differences observed in the results of the present study possibly show that, for the characteristic in question and depending on the cultivars involved in the crosses, inheritance may behave differently with respect to the populations under study, so it is not possible to extrapolate these results to other populations. Furthermore, the method adopted here for seed size measurement was quantitative, expressed in millimeters, while the study by Polle et al. (1941) categorized seed size into classes; that is, they performed a qualitative assessment, disregarding minimal size differences and limiting the population.
The results obtained by means of the predicted gain from selection (Tab. 4) reinforce that, for the improvement of the characteristic under study, the use of simpler breeding methods can be effective, since the gain from the selection was 59% for relatively large seeds and a decrease of just under 30% for relatively small ones. In other words, it is possible to select and recombine individuals that produce large seeds and small seeds, seeking to develop hybrid combinations that produce seeds of intermediate size; this is in addition to the possibility of exploiting these individuals to develop new cultivars for different purposes.
CONCLUSIONS
In the present work, inheritance of the smallest seed size trait in the studied populations is controlled by two genes with incomplete dominance;
Depending on the population under study, the inheritance behavior for the seed size characteristic may behave differently.
Selection of superior individuals for the smallest seed size within an F2 population can occur based on phenotype;
Increased numbers of individuals can be exploited within breeding programs to develop lines or hybrids.