Remark
| 1) Why was this study conducted? |
| Because the encouraging previous results obtained. Which consisted of the generation of rotavirus isolates capable of infecting and efficiently inducing the death of cell lines derived from malignant tumors. These tumor cell-adapted rotaviruses had been selected by in vitro evolution. The identification of the cell surface receptor molecules responsible for the tropism of the rotaviral isolates for these cells was also a motivation to carry out the present research. This study attempted to produce insights about the dependence of oncolytic viruses on cellular receptors to enter the tumor cell since either the receptors seem not to be present in all tumors or they are absent in some cells of the same tumor. Therefore, the identification of more universal receptors such as HSPs is crucial to suggest whether rotaviruses may be candidates for the treatment of breast cancer. |
| 2) What were the most relevant results of the study? |
| The identification of heat shock proteins (HSPs: Hsp90, Hsp70, Hsp60, and Hsp40) on the cell surface, which were required by rotavirus isolates to successfully infect MCF-7 cells. HSPs were found to form complexes with the protein disulfide isomerase (PDI), integrin β3, and heat shock cognate protein 70 (Hsc70) in lipid raft microdomains. |
| 3) What do these results contribute? |
| To strengthening the path of research aimed at finding alternative and complementary strategies for cancer treatment based on oncolytic viruses. These results stimulate further investigation involving the rotavirus isolates tested here as potential oncolytic agents in the context of virotherapy. |
Introduction
The use of virus for cancer treatment dates from the early 20thcentury 1,2. However, virotherapy studies have been undertaken again in the last two decades since oncolytic viruses are a promising tool to be used as an alternative and complementary therapy for treating tumoral entities 3. The use of oncolytic viruses as therapeutic agents is based on their selective replication in tumor cells without being harmful to the patient 4. Oncolytic viruses are characterized by their tumor tropism, their ability to enhance anti-tumor immunity and their ability to be combined with conventional chemotherapy and radiotherapy, and also immunotherapeutic agents 5. Oncolytic viruses within the tumor cells are able to stimulate the immune system facilitating in this way an efficient and durable response to the tumor 6.
To date, several oncolytic viruses have proven to be effective against some tumor cells, including animal models 7-10, pre-clinical studies 11 and clinical trials 11-13. Recent clinical trials have shown the safety, tolerability, and efficacy of some oncolytic viruses used for the treatment of cancers 14. Oncolytic viruses perform multiple interactions with their target tumor cells by functioning as direct cytotoxic agents and therapeutic cancer vaccines to induce cell death through a number of mechanisms including immunogenic apoptosis, pyroptosis, necrosis, and autophagic cell death 14-16. Tumor cell tropism of oncolytic viruses takes advantage of cell surface receptors that facilitate virus binding and penetration into the tumor cells to establish a lytic cycle in tumor cells without affecting normal cells 17. Many viruses have been studied about their oncolytic properties 17 and approximately 40 clinical trials using several oncolytic viruses are being conducted 18. IMLYGICTM(T-VEC/Talimogene Laherparepvec, a genetically engineered herpes simplex virus indicated for local treatment of non-resectable cutaneous, subcutaneous, and nodal lesions in patients with melanoma, is the first FDA-approved oncolytic viral therapy 19.
Cellular heat shock proteins (HSPs) play a central role in maintaining cellular homeostasis and in forming a protective network against stressful challenges, and also in facilitating folding of nascent proteins and refolding of denatures proteins (20. Virus infections are able to induce HSPs which in turn support virus replication. Hsp70, which is particularly induced by virus infections and fever, supports an enhanced expression of virus genes 21. HSPs are expressed at high levels in a wide range of human cancers where they contribute to tumor development by promoting cell proliferation, invasion, metastasis, death, and recognition by the immune system 22,23. Hsp90 has a positive role in the rotavirus infection process in MA104 cells by modulating Akt signaling 24. Heat shock cognate protein (Hsc70) is a receptor used for rotavirus during infection of MA104 cells 25 and is implicated in inducing conformational changes in rotavirus particles 26. Cell surface PDI and integrin (3 are also used as receptors for rotavirus 27,28.
Rotavirus is comprised of three concentric capsid protein layers surrounding a genome composed of 11 double-stranded RNA segments. Rotavirus tropism is crucially dependent on receptor usage and levels of cell surface receptor expression. The growth of rotaviruses in human and monkey cell lines has been shown to be correlated with cell surface expression of some integrins 29. The interaction of rotavirus outer proteins with cell surface receptor molecules mediates its entry into the permissive host cells following some structural changes in the viral outer layer proteins VP4 (VP8* and VP5*) and VP7 30.
Tumor cell-adapted rotavirus isolates have recently been reported which are able to induce cell death in some tumor cells 31. In the present study, we show that these rotavirus isolates are able to successfully infect the human mammary gland adenocarcinoma cell line MCF-7 using some cell surface HSPs as receptors for entry into the cell. Our findings suggest that these rotavirus isolates could be further investigated as a potential therapeutic strategy for treating cancer.
Materials and Methods
Cells and viruses
Human mammary gland adenocarcinoma cell line MCF-7 (ATCC, Cat. No. HTB-22) was propagated in Dulbecco's Modified Eagle's medium (DMEM) (Sigma-Aldrich, St. Louis, MO, USA, Product No. D6429) supplemented with 10% FBS, 100 μg/mL streptomycin and 100 μg/mL penicillin (Eurobio, Les Ulis, France) at 37º C in a 5% CO2 humidified chamber. This cell line was kindly donated by Dr. C. Parra (Department of Microbiology, Faculty of Medicine, Universidad Nacional de Colombia). Rotavirus isolates WT1-5, TRUYO, WWM, WTEW or ECwt-o were generated as previously described 31. Viruses were propagated in MCF-7 cells.
Virus purification
Rotavirus particles were purified as previously reported 32. Briefly, infected cells in Tris-buffered saline (TBS) (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2 and 5 mM CaCl2) were broken by freezing and thawing twice and the lysate was emulsified with 1,1,2-trichlorotrifluoroethane (Freon) and centrifuged at 12 g for 10 min at 4º C. The organic phase was reextracted with TBS and the aqueous fractions were subjected to centrifugation at 120,000 g for 1.5 h at 4º C. The sedimented virus was resuspended in TNC buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM CaCl2) and further centrifuged through a discontinuous sucrose-cesium chloride gradient at 210,000 g for 1 h at 4º C. The viral bands (TLPs and DLPs) were separately collected and subjected to the second cycle of high-speed sedimentation and sucrose-cesium chloride gradient centrifugation. The purified TLPs and DLPs in TNC buffer were stored at - 70º C until used.
Virus infections
MCF-7 cells (5 × 104) in 50 µL DMEM per well were infected at 0.8 MOI with cesium chloride gradient-purified rotavirus isolates WT1-5, TRUYO, WWM, WTEW or ECwt-o that had previously been activated with trypsin (1 µg/mL). The 96-well culture plates were incubated at 37 ºC and the cells were analyzed by an immunocytochemistry assay at 12 h.p.i. Virus titers were determined by infecting cells at 0.04 MOI followed by detection of the percentage of infected cells at 12 h.p.i using an immunocytochemistry assay.
Immunocytochemistry assay
Cells were cultured onto xylol-washed coverslips in 96-well culture plates and fixed at room temperature (RT) with 4% paraformaldehyde (PFA) in PBS at 12 h.p.i. The PBS-washed cells were reacted with rabbit polyclonal antibodies to rotavirus structural proteins (SP) and incubated at 37º C for 1 h. The cells were washed twice with PBS and incubated with HRP-conjugated goat anti-rabbit antibodies (0.133 μg/mL, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) at 37º C for 1 h. After three washes with PBS, rotavirus antigen-positive cells were visualized using 3-amino-9-ethylcarbazole (AEC) substrate (0.64 mg/mL) in 50 mM sodium acetate buffer, pH 5.0, containing 0.04% H2O2. Control cells consisted of non-infected cells or infected cells treated with non-related antibodies. Ten representative images were photographically recorded using a light microscope (VanGuard, Scottsdale, USA) and the mean percentage of infected cells was determined.
Cell viability
MCF-7 cell monolayers were treated with 0.25% trypan blue solution for 1 min at RT. PBS-washed cells were assessed for the exclusion of trypan blue. This assay was used to determine the maximum concentrations of the different antibodies, inhibitors, and proteins that could be used without affecting cell viability. The cells were treated separately with recombinant proteins (rHsp90 and rHsp70) and antibodies against HSPs (90, 70, 60 and 40) and Hsc70, and also with the Hsp90 inhibitors 17AAG 33, NVP-AUY922 34 and radicicol 35, and the Hsp70 inhibitors VER-155008 36 and quercetin 37. Incubation with treatments was carried out for 12 h at 37º C and non-treated cells were used as a control.
Cell membrane isolation
Previously PBS-washed cells (1 x 108) were suspended in hypotonic buffer (5 mM HEPES, 7.4, and 50 mM sucrose) and lysed using a glass Dounce homogenizer for 5 min on ice. The homogenate was added with CaCl2 (10 mM final concentration) and then centrifuged at 3,500 g for 5 min at 4º C. The supernatant was further centrifuged at 20,000 g for 30 min at 4º C. The pellet was resuspended in PBS containing 0.02% sodium azide and stored at 4º C until used within 24 h.
Cell membrane protein isolation
Membrane-associated proteins were solubilized from cell membrane fractions by treating them with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS) for 30 min at 37° C and the solubilized proteins were quantified using a NanoDrop 2.000c spectrophotometer.
ELISA
Solubilized proteins from cell membranes were added to the wells of ELISA plates coated with rabbit antibodies against β3, PDI or Hsc70 and incubated at 4º C overnight, followed by blocking with 0.6% casein for 1 h at 37º C. Plate wells were washed three times with PBS containing 0.05% Tween 20 (PBS-T) for 5 min each. Primary goat antibodies (0.2 µg/mL, Santa Cruz Biotechnology Inc.) against Hsp70, Hsp60 or Hsp40 were separately added to wells and incubated for 1 h at 37º C. After three washes with PBS-T, secondary HRP-conjugated donkey anti-goat antibody (0.133 µg/mL, Santa Cruz Biotechnology Inc. SC-2020) was added, followed by incubation for 1 h at 37º C. The reaction was revealed using o-phenylenediamine dihydrochloride (OPD, PIERCE®) substrate in a stable peroxide substrate buffer. The reaction was stopped with 3N H2SO4 and the absorbance at 492 nm measured in an ELISA plate reader. Results were expressed as delta OD obtained by subtracting the absorbance values of the control wells in which no solubilized protein was added, but antibody treatments were identical. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
In a parallel experiment, cell membrane fractions were separately incubated with rotavirus isolates WTEW, TRUYO, WWM, Wt1-5, or ECwt-o for 1 h at 37º C, followed by solubilization in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150mM NaCl, 1% NP-40, 0.5% DOC and 0.1% SDS) for 30 min at 37º C. Solubilized cell membrane fractions were added to ELISA plates coated with guinea pig antibodies to rotavirus structural proteins and incubated for 1 h at 37º C. After blocking as indicated above and washing three times with PBS-T, the plates were incubated for 1 h at 37º C with primary goat antibodies (0.2 µg/mL, Santa Cruz Biotechnology Inc.) to Hsp90, Hsp70, Hsp60, Hsp40, PDI and integrin β3. After three washes with PBS-T, secondary HRP-conjugated donkey anti-goat IgG (0.133 µg/mL, Santa Cruz Biotechnology Inc. SC-2020) was added and incubated for 1 h at 37º C. Following three times washing with PBS-T, the reaction was revealed with OPD and quantified as indicated above. In the control wells, virus treatment was omitted while other treatments were kept identical.
Lipid raft assay
Cells (1 x 108) in FBS-free DMEM were treated or left untreated with methyl-β-cyclodextrin (10 mM) for 1 h at 37° C and then washed with PBS and lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 2 mM DTT and 1% Triton X-100) in the presence of 2 mM PMSF at 4° C for 20 min. The lysate was added with one volume of 80% sucrose in lysis buffer, placed in the bottom of a centrifuge tube and overlaid with a linear sucrose gradient (30 to 5%). After centrifugation at 120,000 g for 4 h at 4° C, fractions (500 µL) were collected from the top of the gradient and the fractions containing the rafts were treated with 1% Triton X-100 for 30 min at 37° C.
The solubilized fractions were applied to ELISA plates coated with rabbit polyclonal antibodies against integrin β3, blocked with 0.6% casein and then incubated overnight at 4° C. After washing three times with PBS-T, the plate wells were treated with primary goat antibodies (0.2 µg/mL, Santa Cruz Biotechnology Inc.) against Hsp90, Hsp70, Hsp60 or Hsp40 for 1 h at 37º C. Following three times washing with PBS-T, secondary HRP-conjugated donkey anti-goat IgG (0.133 µg/mL, Santa Cruz Biotechnology Inc. SC-2020) was added, followed by incubation for 1 h at 37º C. After PBS-T washing three times, OPD was used to visualize and quantify the reaction as indicated above. Wells, without the addition of solubilized fraction, were used as control wells. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Immunoprecipitation
MCF-7 cell membrane-enriched fractions were separately mixed with trypsin-activated rotavirus isolates WTEW, TRUYO, WWM, Wt1-5 or ECwt-o. As a control, MA104 cell membrane-enriched fractions were mixed with trypsin-activated rotavirus isolates Wt1-5. After incubation for 45 min at 37° C, RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% DOC and 0.1% SDS) was added and the preparation incubated for 30 min at 37 °C. The solubilized membranes were mixed with protein G agarose beads that had been coated with rabbit anti-rotavirus and blocked with casein, followed by constant agitation for 30 min at RT. The PBS washed beads were sedimented and treated with Laemmli’s sample buffer for 3 min in a boiling water bath. The denaturalized proteins were analyzed by SDS-PAGE/Western blotting. The casein-blocked membranes were incubated with a mix of goat antibodies (0.2 μg/mL each, Santa Cruz Biotechnology Inc.) against Hsp90, Hsp70, Hsp60 and Hsp40 or alternatively with rabbit antibodies against rotavirus SP for 1 h at 37° C. In the case of MA104 cells with Wt1-5, antibodies against integrin β3, Hsc70, PDI or HSPs (90, 70, 60, 40) were used in the SDS-PAGE/Western blotting analysis. Membranes were washed three times with PBS-T and incubated with secondary HRP-conjugated donkey anti-goat IgG or goat anti-rabbit antibodies (0.2 μg/mL, Santa Cruz Biotechnology Inc.) for 1 h at 37° C. After PBS-T washing, the membrane was revealed with Western blotting Luminol reagent (Santa Cruz Biotechnology Inc. SC-2048). Solubilized cell membrane fractions treated with protein G agarose beads without antibody were used as a control.
Interactions of HSPs (Hsp90, Hsp70, Hsp60 y Hsp40) with rotavirus isolates was tested using biotinylation of cell surface coupled with immunoprecipitation of receptors for rotavirus virions. PBS-washed cells (1 x 107) were treated with 0.5 mg/ml of EZ-Link Sulfo-NHS-SS-Biotin (sulfosuccinimidyl-2-[biotinamido] ethyl-1, 3-dithiopropionate) for 30 min at 4° C. Cells were washed with PBS containing 100 mM glycine for 30 min at 4° C and then separately inoculated with trypsin-activated rotavirus isolates WTEW, TRUYO, WWM, Wt1-5 or ECwt-o at MOI 4 for 30 min at 4° C. Unbound rotaviruses were removed by washing the cells twice with PBS before treatment with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% DOC and 0.1% SDS) for 30 min at 37° C. The cell lysate was mixed with protein G agarose beads (Bio- Rad®, 90-180 μm) that had been coated with rabbit antibodies to rotavirus and blocked with 0.5% casein and incubated for 30 min at RT with constant agitation. The beads were washed three times with PBS and the pellet was treated with Laemmli’s sample buffer for 3 min in a boiling water bath. The solubilized proteins were analyzed by SDS-PAGE/Western blotting. Proteins were electrophoretically transferred to a PVDF membrane using 10 mM CAPS buffer, pH 10.5, and the membrane blocked with 5% casein. The membrane was reacted with streptavidin-HRP for 1 h at 37° C and the reaction visualized with Western blotting Luminol reagent. The cell lysate containing rotavirus and incubated with protein G agarose beads without anti-rotavirus antibodies were used as a control. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
FACS analysis
PBS-washed MCF-7cells were further washed with PBS containing 1 mM EDTA for additional 5 min. The cells were fixed in 4% paraformaldehyde (PFA) in PBS at 37° C for 30 min and then washed twice with PBS. Expression of cell surface proteins Hsc70, Hsp90, Hsp70, Hsp60, Hsp40, PDI, and integrin β3 was detected by incubating separately the cells with primary goat antibodies (0.2 µg/mL, Santa Cruz Biotechnology Inc.) to each cellular protein for 45 min at 4° C. Goat irrelevant antibody (IgG2a) was used as an isotype IgG control. After washing twice with PBS containing 1% BSA, the cells were incubated with donkey anti-goat IgG coupled to Alexa Fluor488 (0.4 µg/mL, Santa Cruz Biotechnology Inc. SC-362255) for 20 min at 4° C. After washing with PBS, the cells were analyzed using a Dako CyAn flow cytometer (Dako Cytomation). Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Antibody blocking assay with F(ab')2 fragments
MCF-7 cells (1 x 107) were washed with RPMI medium without FBS and then separately treated with rabbit F(ab')2 fragments, generated by pepsin digestion in our laboratory, against 20-mer synthetic peptides derived from Hsp90, Hsp70, Hsp60, and Hsp40 sequences. (The peptide sequences were as follows: Hsp90 (620-RDNSTMGYMAAKKHLEINPDHS-641); Hsp70 (374) (705-QIQQYMKIISSFKNKEDQYDHLD-727); Hsp70 (375) (646-NSFTLKLEDTENWLYEDGDQPKQ-668); Hsp70 (376) (741-AMEWMNNKLNLQNKQSLTMDP-761); Hsp60 (393-RLAKLSDGVAVLKVGGTSDVEVN-415); Hsp40 (251-GSDVIYPARISLREALCGCTVNV-273). F(ab')2 fragments were used at dilutions 1:40, 1:80, 1:160, 1:320; 1:640 and 1:1280 and incubated for 1 h at 37° C. Unbound F(ab')2 fragments were removed by resuspending cells in 1 ml of cold PBS before centrifugation at 600 g for 5 min and removal of the supernatant. Cells were separately inoculated with 100 µL of cold RPMI medium containing rotavirus isolates WTEW, TRUYO, WWM, Wt1-5 or ECwt-o at MOI 2 and incubated for 45 min at 4° C. The inoculum was removed, and cells were washed three times with PBS and lysed with RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS and 5 mM PMSF) for 1 h at 37° C.
The lysate was applied in a 96-well ELISA plate coated with guinea pig antibodies to rotavirus structural proteins and blocked with 0.6% casein before incubation for 1 h at 37° C. After three washes with PBS-T, goat antibodies to rotavirus SP were added, and then incubated for 1 h at 37° C before three washes with PBS-T. Secondary HRP-conjugated donkey anti-goat antibody (0.133 μg/mL, Santa Cruz Biotechnology Inc. SC-2020) was added, followed by incubation for 1 h at 37° C. The reaction was visualized with OPD as indicated above. Cells without F(ab')2 fragment pre-treatment and inoculated with rotavirus isolates were used as a positive control, whereas ovalbumin instead of rotavirus was used as a negative control. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Viral overlay protein binding assay (VOPBA)
MCF-7 cells (1 x 108) were lysed using Laemmli’s sample buffer (0.125 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 5% 2-ME, 0.02% bromophenol blue) and the solubilized proteins subjected to SDS-PAGE (10%) before being electrophoretically transferred to PVDF membrane (Merck Millipore, Billerica, MA, USA) using 10 mM CAPS buffer, pH 10.5. The 5% casein-blocked membrane was cut into two lanes (1 and 2) and one of them (1) incubated with trypsin-activated (10 µg/mL) WTEW TLPs in FBS-free medium overnight at 4° C. This membrane lane was immediately washed with PBS and incubated with 2.5% glutaraldehyde during 30 min at 37° C before being washed three times with PBS and incubated with rabbit anti-rotavirus antibodies. The membrane lane 2 was incubated with goat antibodies against Hsp90, Hsp70, Hsp60, and Hsp40 (Santa Cruz Biotechnology Inc.). After both membrane lanes (1 and 2) were separately incubated for 1 h at 37° C and washed twice with PBS, they were respectively incubated with secondary HRP-conjugated goat anti-rabbit IgG or HRP-conjugated donkey anti-goat IgG for 1 h at 37° C. The PBS-washed membrane lanes were developed with Western blotting Luminol reagent. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Blocking assay with antibodies to HSPs
MCF-7 cell monolayers (80% confluence) in 96-well culture plates were separately incubated with antibodies raised in rabbits against a 20-mer synthetic peptide derived from Hsp90 (620-641), synthetic peptides derived from three Hsp70 domains (Hsp70 [705-727], Hsp70 [646-668] and Hsp70 [741-761]), Hsp60 [393-415]) or Hsp40 [251-273] at dilutions of 1:40, 1:80 and 1:160. After incubation for 1 h at 37° C, the cells were washed with FBS-free RPMI medium and placed on ice for 15 min. The cells were separately inoculated with the rotavirus isolates WTEW, TRUYO, WWM, Wt1-5 or ECwt-o at MOI 0.8 for 45 min at 4° C. Unbound virus was removed by washing twice with PBS and the cells incubated for 12 h at 37° C, before the detection of cells being positive to rotavirus antigens using the immunocytochemistry assay described above. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Inhibition treatment of Hsp90 and Hsp70
Cells at 80% confluence were incubated with Hsp90 inhibitors 17AAG (10, 20, 40 and 80 nM) (Kao, 2016), NVP-AUY922 (2, 4, 8 and 16 nM) [34] or radicicol (7.5, 25, 50, and 100 nM) 35, and Hsp70 inhibitors VER-155008 (10, 20, 40 and 80 nM) 36 or quercetin (75, 150, 300 and 600 µM) 37 for 1 h at 37 ºC. Cells were separately inoculated with WWM, TRUYO, Wt1-5, WTEW or ECwt-o at MOI 0.8, for 45 min at 4º C. The DMEM-washed cells were further incubated for 12 h at 37º C and then fixed with PFA before submitting them to immunocytochemistry analysis. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Expression of recombinant HSPs in E. coli
Expression constructs (pRSET-His-HSP90α wt, HSP90α-PGEXgp2, PMS-HSP70 AMP and PET-16b-HSP70) were kindly donated by Dr. Y. Miyata (Department of Cell & Developmental Biology, Graduate School Biostudies, Kyoto University). Aliquots from bacterial competent cells (DH5α or BL21 (DE3)) in LB medium (200 µl) containing ampicillin (100 µg/ml) were mixed with plasmid DNA (10 ng) in nuclease-free solution (100 µL) (100 mM KCl, 30 mM CaCl2 and 50 mM MgCl2) with gentle agitation before being incubated on ice for 20 min. A 42º C heat-shock of 2 min was performed followed by immediate incubation on ice for 2 min. The cells were inoculated into LB medium (5 mL) containing ampicillin (100 µg/mL) before incubation at 37º C until the exponential growth phase was reached. Competent bacterial cells without addition of plasmid DNA but subjected to all the same transfection steps were used as a control. Invitrogen™ Purelink™ Quick Plasmid Miniprep kit (Cat # K2100-10) was used to purified DNA from bacterial cells as directed by the manufacturer.
Bacterial cells (DH5α or BL21 (DE3)) harboring the plasmids were subjected to induction to express recombinant proteins by culturing them in LB medium (25 mL) containing ampicillin (100 µg/mL) and 3% ethanol for 12 h at 37º C with constant agitation (150 rpm) 38. Non-transfected cells and non-induced transfected cells were used as controls. Aliquots (5 ml) of bacterial culture were inoculated into LB medium (500 mL) and further incubated at 37º C with constant agitation (150 rpm) until the exponential growth phase was reached. To express recombinant proteins, 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added for 4 h at 37º C with constant agitation (150 rpm). The cell suspension was subjected to centrifugation at 12,000 g for 10 min and the sedimented bacterial cells were resuspended in PBS containing 1% N-lauryl sarcosine and 100 µg/mL lysozyme. After incubation for 30 min at 4º C at constant agitation, the cell lysate was sonicated at 45% amplitude for 5 cycles of 30-sec pulses with 60-sec pauses. The cell lysate was centrifuged at 12,000 g for 10 min at 4° C and the soluble proteins in the supernatant were collected.
The proteins present in the 12,000 g supernatant were concentrated by adding polyethylene glycol (PEG) 8.000 (10% final concentration), followed by constant agitation for 12 h at RT. After centrifugation at 12,000 g for 30 min at 4° C, the supernatant was discarded, and an aliquot of precipitated proteins was resuspended in Laemmli’s sample buffer and heated in a boiling water bath for 3 min. The expression of recombinant proteins was analyzed by SDS-PAGE/Western blotting applying the same amounts of total protein into each gel well. The membrane was treated with primary antibodies against Hsp90 or Hsp70. Secondary HRP-conjugated donkey anti-goat IgG (0.4 (g/mL, Santa Cruz Inc. SC-2020). The reaction was visualized using Western blotting Luminol reagent. Lysates from non-transfected bacterial cells (DH5α or BL21 (DE3)) and non-induced transfected BL21 (DE3) cells were used as controls. The remaining PEG-precipitated protein was resuspended in PBS containing 14.2 nM 2-ME and spectrophotometrically measured at 280 nm against a BSA calibration curve.
HSP pre-treatment of virus particles
Virus particles of rotavirus isolates TRUYO, WWM, WTEW, WT-15, and ECwt-o were treated with HSPs by mixing virions with purified recombinant proteins Hsp70 or Hsp90 at concentrations of 100, 50, 25, 12.5, 6.25 and 3.125 ng/mL for 24 at 4º C. The HSP-treated virions were inoculated into MCF-7 cells followed by incubation for 12 h at 37º C. Virions treated with BSA (100 ng/ml) or Hsc70 (100 ng/mL) or left without treatment were used as a control. After incubation, the cells were frozen and thawed twice before treatment with RIPA buffer (150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS and 50 mM Tris-HCl, pH 7.5). After incubation for 1 h at 37 ºC, the lysate was applied into ELISA plates coated with guinea pig antibodies to rotavirus SP. Primary goat antibodies to rotavirus particles and secondary HRP-conjugated rabbit anti-goat antibodies were used. The reaction was revealed with OPD substrate in 100 mM citrate buffer, pH 5.0, and read at 492 nm.
Cell transfection and expression analysis
The plasmids Hsp90α-<human>/pFLAG-CMV2, pCDNA3-FLAG Hsp90α, pCDNA3.1 (+) Hsp70, and pCMV-Hsp70 were transfected into MCF-7 cell monolayers (85% confluence). Plasmids (0.1 ng/mL) in RPMI medium containing 10% FBS (Gibco®), 20 U/ml penicillin, 40 U/ml streptomycin (Invitrogen®) and 6 μg/ml Hexadimethrine bromide (Polybrene®, SIGMA Cas Number 2878-55-4) were separately added to cells. After 48-h culture at 37 °C in a humidified chamber with 5% CO2, the cells were fixed with methanol for 45 min at RT and washed with PBS. The cells were reacted with either with antibodies raised in rabbits against a 20-mer synthetic peptide derived from Hsp70 or mAb (143 µg/mL, Santa Cruz Biotechnology Inc.) to the Hsp90 sequence motif DYKDDD (FLAG). After three PBS washes, the cells were incubated with secondary FITC-conjugated bovine anti-rabbit antibodies (0.8 µg/mL, Santa Cruz Biotechnology Inc. SC-2365) or secondary FITC-conjugated goat anti-mouse antibodies (0.8 µg/mL, Santa Cruz Biotechnology Inc. SC-2010). Cells expressing rHsp70 or rHsp90 were separately inoculated with TRUYO, WWM, WTEW, WT-15 or ECwt-o at 24 h post-transfection. Inoculated cells were cultured for 12 h at 37° C. Non-transfected or transfected cells with empty vectors were infected with rotavirus isolates and used as controls. Virus antigen-positive cells were analyzed by immunocytochemistry staining and immunofluorescence microscopy. Data were presented as mean ( standard deviation from three independent experiments performed in duplicate.
Aliquots of non-transfected or transfected cells were washed with PBS and treated with Hsp90 inhibitor 17AAG (80 nM) for 1 h at 37º C. The cells were inoculated with WWM, ECwt-o, TRUYO, Wt1-5 or WTEW, at MOI 0.8 and incubated for 45 min at 4 ºC, before incubation for 12 to 14 h at 37º C. The cells were fixed with cold methanol and washed with PBS, prior analysis of infection by immunocytochemistry staining using primary guinea pig anti-rotavirus antibodies and secondary HRP-conjugated goat anti-guinea pig IgG (0.133 µg/mL, Santa Cruz Biotechnology Inc. SC-2438) as indicated above. The cells were washed twice with PBS and the reaction developed with 3-amino-9-ethylcarbazole (AEC) substrate in 50 mM Na-acetate buffer, pH 5.0, and 0.03% H2O2. The same AEC-developed coverslips were washed with PBS and tested for the expression of rHsp90 y rHsp70 in the transfected cells. The cells were reacted with the rabbit antibodies against the Hsp70 synthetic peptide or the mAb to the Hsp90 sequence motif DYKDDDDK (FLAG). Following three washes with PBS, the cells were incubated with the secondary FITC-conjugated goat anti-guinea pig IgG or secondary FITC-conjugated rabbit anti-mouse IgG. Ten representative photographs were taken.
Statistical analysis
The data obtained were analyzed using the GraphPad Prism v5.0® program. A two-way ANOVA test was used to determine the statistically significant differences between the means of each treatment. The level of significance was plotted according to the following conventions: * = p <0.1 (Trend); ** = p <0.05 (significant difference); *** = p <0.01 (highly significant difference) and ns = non-significant differences.
Results
Expression of HSPs, PDI and integrin β3 on the cell surface
The flow cytometry analysis showed that the percentage of positive cells for Hsp70, Hsp90, Hsp60 and Hsp40, Hsc70, PDI and integrin β3 was 83.5%, 51.1%, 58.4%, 67.6%, 75.33%, 35.8% and 47.18%respectively. No fluorescent signals were detected for control cells consisting of non-antibody-treated cells, only secondary antibody-treated cells or non-related antibody-treated cells (Fig. 1A).
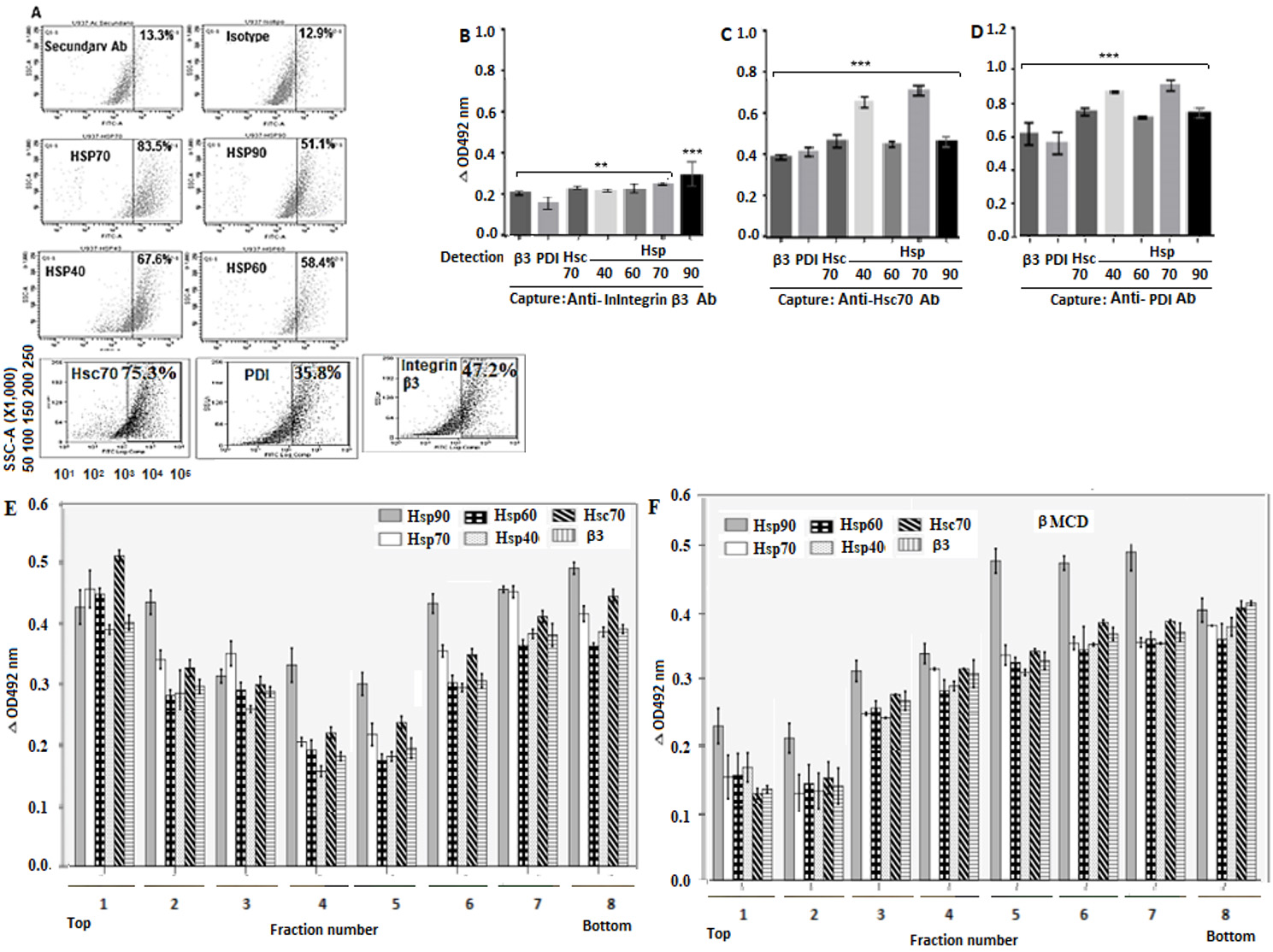
Figure 1 Localization of Hsp90, Hsp70, Hsp60, Hsp40, PDI and integrin β3 in cell membrane. A. Cells were fixed in 4% PFD and incubated with goat primary antibodies against the above-mentioned proteins. Alexa Fluor488-conjugate donkey anti-goat IgG was used as secondary antibody. Immunofluorescence analysis was performed in duplicate using a Cyan (Dako) flow cytometer and a FlowJo software. B-D. RIPA buffer-solubilized cell membrane-enriched fractions were separately added to ELISA plates coated with rabbit capture antibodies against Hsc70, PDI or integrin β3. Goat primary antibodies to Hsp90, Hsp70, Hsp60, Hsp40 Hsc70, PDI or integrin β3 were added for detection before addition of secondary HRP-conjugated donkey anti-goat antibody. Plates were read at 492 nm and the results expressed as delta OD after subtracting control values. E, F. Cells with (E) or without (F) methyl- β-cyclodextrin treatment were lysed and subjected to sucrose gradient centrifugation before collection of gradient fractions containing rafts. The RIPA buffer-solubilized fractions were added to ELISA plates coated with rabbit antibodies to integrin β3. Goat primary antibodies to Hsp90, Hsp70, Hsp60, Hsp40 or integrin β3 were added and then secondary HRP-conjugated donkey anti-goat IgG. The reaction was visualized using OPD and the plates read at 492 nm. Data are shown as means ( standard deviation from three independent experiments performed in duplicate.
Interactions of HSPs with PDI, integrin β3 and Hsc70
After detection with secondary antibodies, the mean delta absorbance values indicated that the captured integrin β3 was able to interact with PDI, Hsc70, Hsp40, Hsp60, Hsp70 and Hsp90 (Fig. 1B). The captured Hsc70 interacted with integrin β3, PDI, Hsp40, Hsp60, Hsp70, and Hsp90 (Fig. 1C). The captured PDI interacted with integrin β3, Hsc70, Hsp40, Hsp60, Hsp70 and Hsp90 (Fig. 1D). The ELISA results suggest that the cellular proteins tested are clustered probably forming a complex but details of the interactions, whether direct or indirect, are not clear from this assay.
Lipid raft microdomain-associated HSPs
After detection with antibodies to Hsp90, Hsp70, Hsp60, Hsp40, Hsc70, and integrin β3, the low-density fractions (2 to 7) from cells without methyl-β-cyclodextrin treatment showed relatively higher delta absorbance values for these proteins than those shown for high-density fractions (Fig. 1E). In the case of methyl-β-cyclodextrin-treated cells, fractions 5, 6, 7 and 8 showed relatively higher delta absorbance values for the tested proteins than those found for low-density fractions (Fig. 1E).
Interaction of rotavirus WTEW, TRUYO, WWM, Wt1-5 and ECwt-o with Hsp90, Hsp70, Hsp60, Hsp40, integrin β3, PDI and Hsc70
The analysis of antibody reactions for the assays using cell membrane-enriched fractions showed that all the cellular proteins tested were able to interact with all the rotavirus isolates studied, according to the delta absorbance values relative to the control consisting of the corresponding virus suspension in RIPA buffer instead of RIPA lysates from virus-inoculated cells (Fig. 2A-E). Although differential delta absorbance values were found for some particular rotavirus isolate-protein interactions, Hsp70 showed the highest delta absorbance values (Fig. 2A-E). Similarly, positive and significant interactions were observed between rotavirus isolates and the cellular proteins studied when cells were used instead of cell membrane-enriched fractions (Fig. 2F-J).
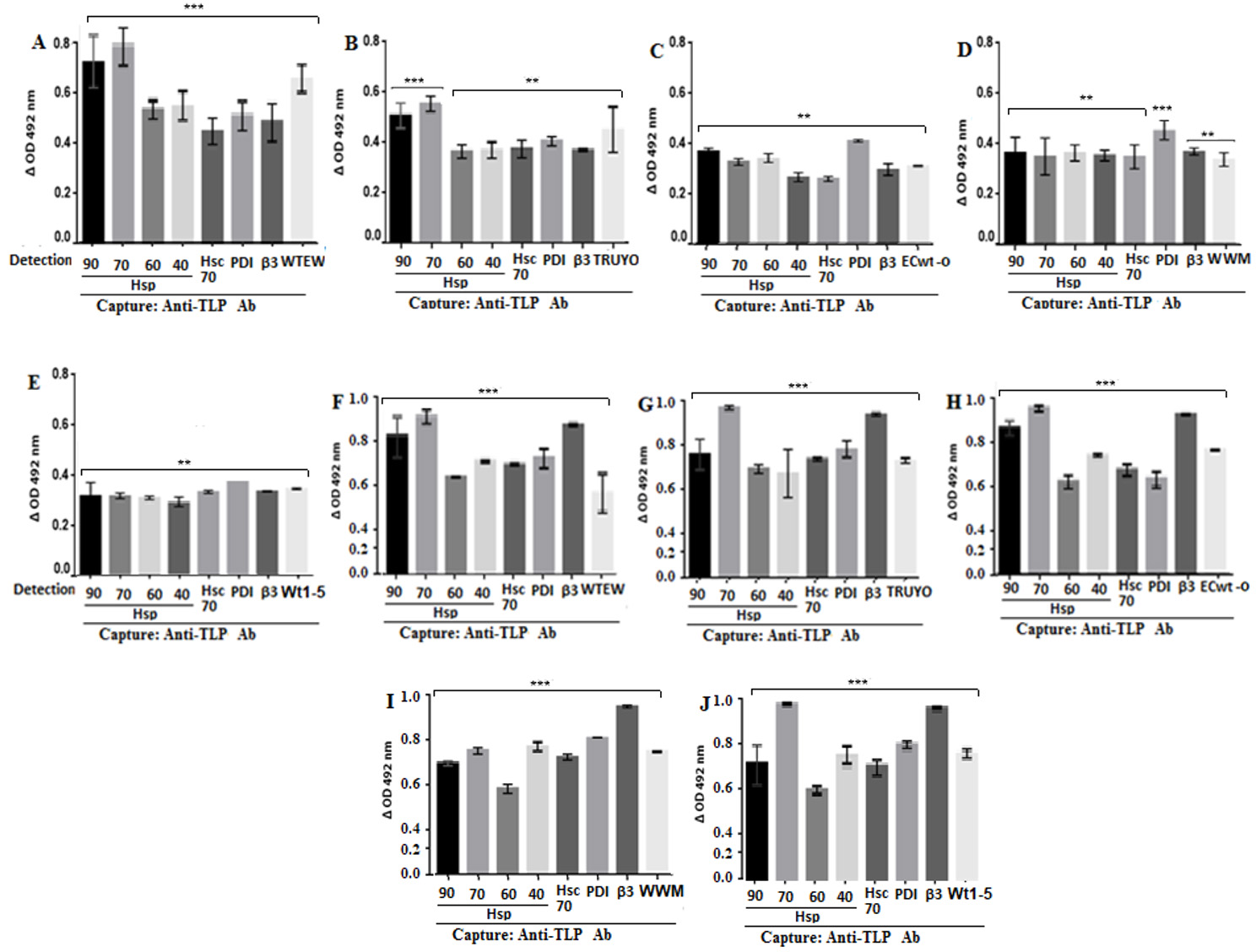
Figure 2 Binding of rotavirus isolates to Hsp90, Hsp70, Hsp60, Hsp40, Hsc70, PDI and integrin β3. MCF-7 cells (A-E) or their cell membrane enriched fractions (F-J) were incubated with the indicated rotavirus isolates. After PBS-washing, the preparations were RIPA-solubilized and applied into ELISA plates coated with rabbit capture antibodies to rotavirus structural proteins. For detection, goat antibodies to HSPs, PDI, Hsc70, integrin β3 or to rotavirus were used and the reaction measured using secondary HRP-conjugated donkey anti-goat IgG and OPD substrate.
Inhibition of rotavirus binding to the cell surface by F(ab')2 fragments against HSPs
After the excess of the virus was washed out, the capture ELISA analysis of RIPA cells lysates revealed that the F(ab')2 fragment treatment decreased rotavirus antigens in a concentration-dependent manner (Fig. 3A-E). All the F(ab’)2 fragment used produced comparable inhibitory effects on all five rotavirus isolates tested.
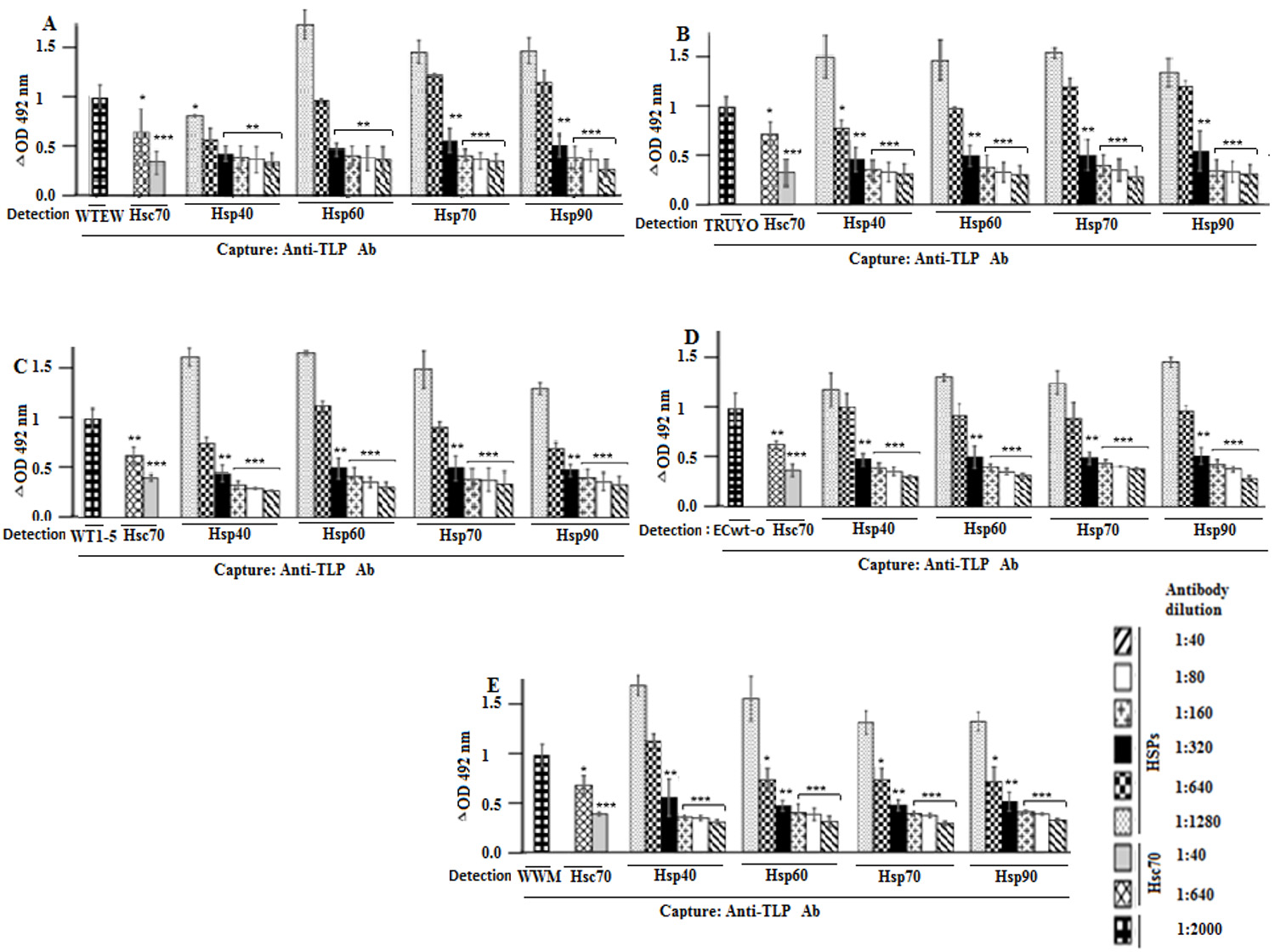
Figure 3 Blocking of rotavirus binding to cell surface by antibodies against Hsp90, Hsp70, Hsp60, Hsp40, and Hsc70. A-F. MCF-7 cells were incubated with F(ab’)2 antibody fragments against to Hsp90, Hsp70, Hsc70, Hsp60 or Hsp40 at the indicated dilutions, followed by incubation with the rotavirus isolates indicated in panels. After PBS- washing, the cells were solubilized with RIPA buffer before addition to ELISA wells coated with guinea pig capture antibodies to rotavirus particles. For detection, rabbit antibodies to rotavirus particles were used and the reaction measured using secondary HRP-conjugated donkey anti-rabbit IgG and OPD substrate. Data are means values ( standard deviation from three independent experiments performed in duplicate.
Co-immunoprecipitation of HSPs and structural antigens of rotavirus isolates
The SDS-PAGE/Western blotting analysis of the co-immunoprecipitated proteins showed four reactive bands corresponding to Hsp90, Hsp70, Hsp60 and Hsp40 (Fig. 4A). These results suggest that the rotavirus isolates studied were able to bind to these HSPs. These results were confirmed by biotinylation of cell surface proteins before SDS-PAGE/Western blotting analysis of the separated HSPs (Anex 1). The co-immunoprecipitation analysis of MA104 cells incubated with Wt1-5 showed that only integrin β3, Hsc70, and PDI co-immunoprecipitated, but not Hsp90, Hsp70, Hsp60 or Hsp40 (Fig. 4B). These results suggest that tumor cell-adapted rotavirus Wt1-5 has gained function since it can interact with these cell surface HSPs.
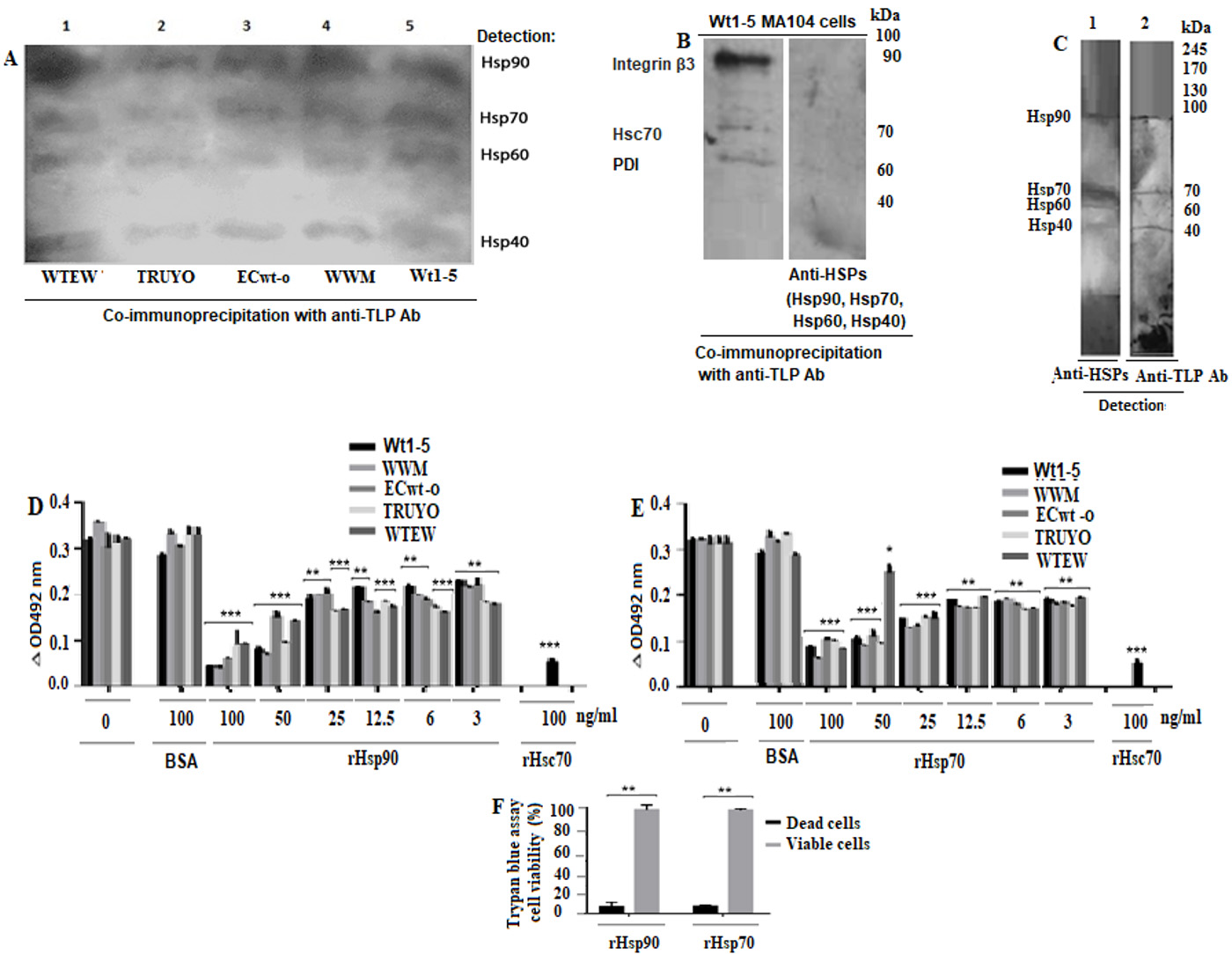
Figure 4 Binging of rotavirus particles to Hsp90, Hsp70, Hsp60 and Hsp40, and inhibition of rotavirus infection by competition for binding to these HSPs. A. MCF-7 cell membrane-enriched fractions were separately incubated with the indicated rotavirus isolates. The solubilized cell membrane fractions were immunoprecipitated with agarose bead-coupled antibodies against rotavirus particles. The immunoprecipitated proteins were analyzed by SDS-PAGE/Western blotting using antibodies against the indicated HSPs and luminol-based detection. B. MA104 cell membrane-enriched fractions were incubated with rotavirus isolate. The solubilized cell membrane fractions were immunoprecipitated with agarose bead-coupled antibodies against rotavirus particles. The immunoprecipitated proteins were analyzed by SDS-PAGE/Western blotting using antibodies against integrin β3, Hsc70, and PDI or HSPs and luminol-based detection. C. A VOBPA assay was conducted by lysing MCF-7 cells with RIPA buffer followed by non-denaturing and non-reducing PAGE and Western blotting. The membrane was incubated with rotavirus WTEW, washed quickly and treated with 2.5% glutaraldehyde. The membrane was treated with a mix of antibodies to the HSPs (lane 1) indicated above or antibodies to rotavirus TLPs (lane 2). Reactions were revealed with Western blotting Luminol Reagent. D, E. Rotavirus isolates (TRUYO, WWM, WTEW, WT-15 and ECwt-o) at MOI 0.8 were treated with the indicated concentrations of rHsp90 (D) or rHsp70 (E) before incubation with MCF-cells. Rotavirus isolates were treated with BSA (100 ng/mL) or Hsc70 (100 ng/mL) or left untreated to be used as controls. The frozen and thawed cells were treated with RIPA buffer and the lysate applied on ELISA plates coated with Guinea pig capture antibodies to rotavirus particles. Detection was performed with rabbit primary antibodies to rotavirus particles. The reaction was revealed with secondary HRP-conjugated goat anti-rabbit IgG and OPD. F. Cell viability of cells treated with recombinant proteins (rHsp90 or rHsp70) (100 (g/mL) was determined using trypan blue exclusion test. Data are presented as means ( standard deviation from three independent experiments performed in duplicate.
VOPBA analysis of rotavirus WTEW on MCF-7 cell proteins
The membrane that was probed with rabbit anti-rotavirus antibodies showed four reactive bands that nearly matched the Hsp90, Hsp70, Hsp60, and Hsp40 bands detected with the respective antibodies in a parallel lane of the Western blotting membrane (Fig. 4C). These findings confirm that rotavirus WTEW interacts with these HSPs.
Inhibition of rotavirus infection by incubation of virions with rHsp90 and rHsp70
Capture ELISA analysis of the cell lysates after incubation for 12 h at 37( C showed that viral antigens were decreased by pre-treatment with the recombinant proteins in a concentration-dependent manner when the respective virus isolate without recombinant protein pre-treatment was used as a control (100%) (Fig. 4D). The lowest inhibition of the viral antigen accumulation produced by rHsp90 was 42.9%, 38.5%, 26.1%, 41.3% and 32.5% for isolates WTEW, TRUYO, ECwt-o, WWM and Wt-15, respectively, whereas the highest inhibition observed was 72%, 79%, 75.5%, 91.2% and 88%, respectively (Fig. 4D). Similar inhibitory effects were found when viral particles were pre-treated with rHsp70. The lowest inhibitory effect ranged from 22.8% to 43.5%, whereas the highest one ranged from 60% to 80% (Fig. 4E). The pre-treatment of viral particles with Hsc70 (100 ng/ml), used as a positive control, reduced viral antigen accumulation by 82.2% for WTEW. Cell viability of MCF-cells treated with rHsp90 and rHsp70 was 96% and 94%, respectively, according to the trypan blue exclusion test (Fig. 4F). These results suggest that the recombinant HSPs assayed stably bind to viral particles preventing them from binding the cell surface counterparts. It is less probable that rHsp90 or Hsp70 binding renders viral particle inactive.
Inhibition of rotavirus infection by antibodies against HSPs
As shown in Fig. 5A-E, antibody treatment provoked a significant decrease in rotavirus infection as determined by immunocytochemistry analysis of the structural rotavirus antigens accumulated at 12 h.p.i. The highest Hsp90 antibody concentration used was able to reduce viral infectivity by 45% (WTEW), 65% (TRUYO), 55% (Wt1-5), 41% (ECwt-o) and 42% (WWM) (Fig. 5A-E). Antibodies to Hsp70 reduced infectivity by 40% (WTEW), 48% (TRUYO), 55% (WT1-5), 55% (ECwt-o) and 56% (WWM) (Fig. 5A-E). Infectivity reduction of 50% (WTEW), 50% (TRUYO), 66% (Wt1-5), 69% (ECwt-o) and 40% (WWM) (Fig. 5A-E) was produced by Hsp60 antibodies, whereas antibodies to Hsp40 reduced infectivity by 60% (WTEW), 67% (TRUYO), 51% (Wt1-5), 40% (ECwt-o) and 32% (WWM). These results suggest that the HSPs tested are involved significantly in the infection of MCF-7 cells with the above-mentioned rotavirus isolates.
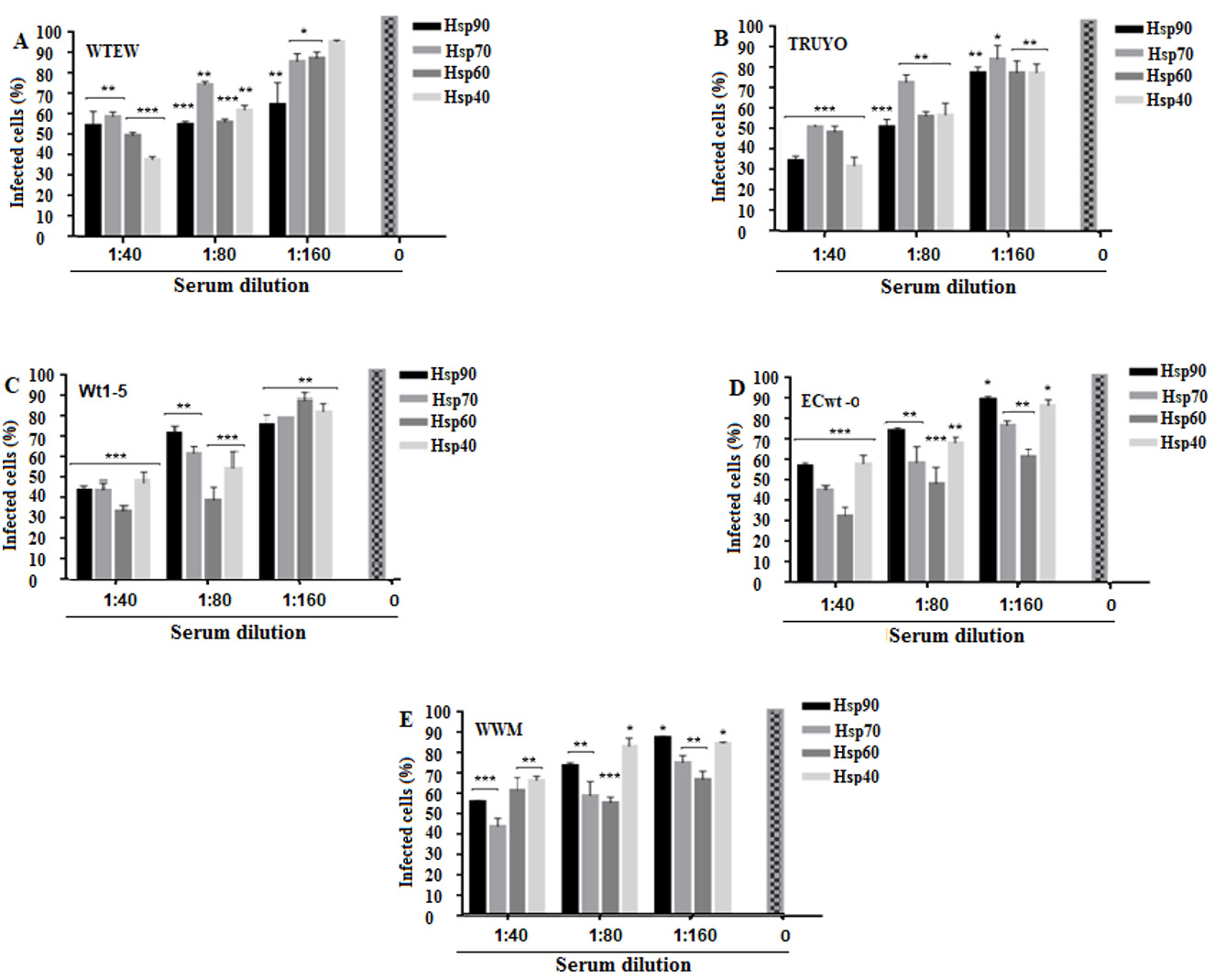
Figure 5 Decrease of rotavirus infection by antibodies to Hsp90, Hsp70, Hsp60 or Hsp40. A-E. MCF-7 cells in culture medium were separately treated with a serial dilution of antibodies to the indicated HSPs for 45 min at 4° C, DMEM-washed and then incubated with the rotavirus isolates indicated in panels. After 12 h.p.i., structural rotavirus antigens were detected by immunocytochemistry assay. Virus inoculated cells without antibody pre-treatment were used as a control. Data were expressed as percentage of infected cells. Data are shown as mean percentages ( standard deviation from three independent experiments performed in duplicate.
Effect of inhibitors of Hsp90 and Hsp70 on rotavirus infection
Pre-incubation with 17AAG at the highest concentration used (80 nM) reduced the percentage of virus infection by 4% to 70%, depending on the virus isolate used (Fig. 6A). NVP-AUY922 reduced viral infection by 60% to 66% and radicicol by 80% to 90%, depending on the rotavirus isolate implicated at the highest concentration of inhibitor used (16 nM and 100 nM, respectively) (Fig. 6B,D). Pre-incubation with VER-15008 led to viral inhibition by 63% to 80%, whereas quercetin inhibited viral infection by 81% to 90%, depending on the rotavirus isolates used when the inhibitors were used at their highest concentrations (75 (M and 600 (M, respectively) (Fig. 6C,E). Pre-incubation of cells with 300 (M quercetin at 43( C for 30 min before infection with WTEW or Wt1-5, led to an inhibition of viral infection by 100% and 80%, respectively, whereas pre-incubation at 37( C for 1 h inhibited virus infection by 70% in both cases (Fig. 6F). The highest concentrations of the inhibitors assayed were selected by testing their effects on cell viability. Cell viability, as determined by the trypan blue exclusion test, was 99.5%, 98.6%, 99.5%, 494.1% and 90% for 17AAG, NVP-AUY922, quercetin, radicicol and VER-155008, respectively (Fig. 6G). Overall, these results are consistent with the participation of Hsp90 and Hsp70 in the infection process of rotavirus in MCF-7 cells.
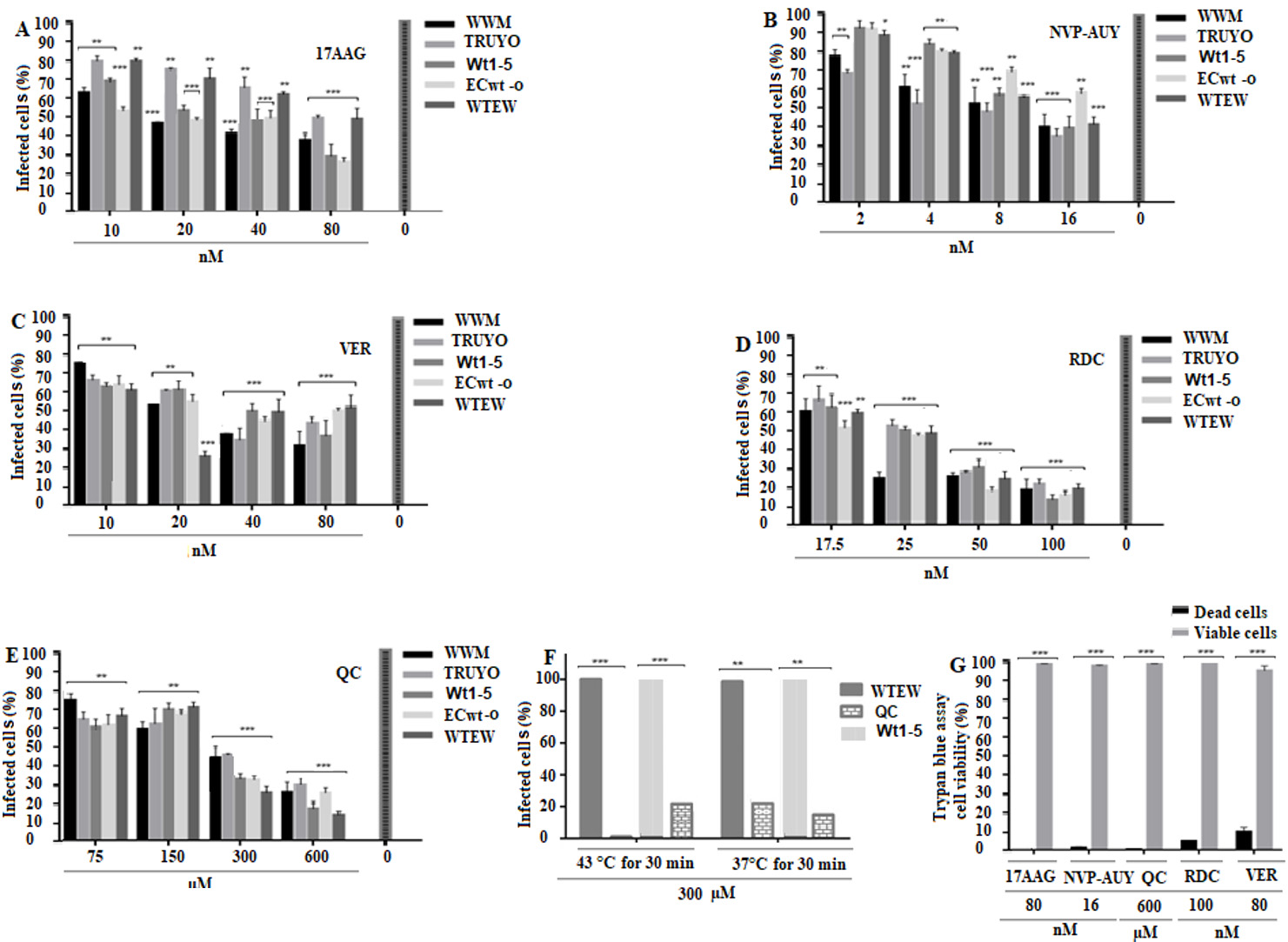
Figure 6 Effect of inhibition of Hsp90 and Hsp70 on rotavirus infection. A-E. MCF-7cells were seeded in 96-well plates with different concentrations of Hsp90 inhibitors (17AAG, NvP Auy or Radicicol) or Hsp70 inhibitors (Quercetin (QC) or VER155008) for 1 h at 37° C. Cells were DMEM-washed and inoculated with the rotavirus isolates indicated in panels and incubated for 45 min at 4° C.Cells were DMEM-washed and inoculated with the rotavirus isolates indicated in panels and incubated for 12 h at 37( C. Viral structural antigens were detected by immunocytochemistry assay and infection was expressed as percentage of infected cells. F. Cells were incubated with 300 (M QC for 30 min at 43( C or 37( C and the percentage of infected cells determined by immunocytochemistry assay using rabbit antibodies to rotavirus structural antigens. G. Trypan blue-cell viability assay of cells treated with 600 (M of the inhibitors indicated in the bottom of the panel. Data are shown as mean percentages ( standard deviation from three independent experiments performed in duplicate.
Effect of rHsp90 and rHsp70 overexpression on rotavirus infection
The transfected cells showed the respective positive signals for either recombinant protein (Fig. 7 A-7D ) while non-transfected cells showed negative fluorescence (Fig. 7 E ). The percentages of infected cells without transfection and without inhibitor treatment were used as a control. These percentages of infection were 90%, 82%, 61%, 80% and 70% for WWM, TRUYO, Wt1-5, ECwt-o and WTEW, respectively, whereas these percentages were increased to 95%, 95%, 70%, 96%, and 83%, respectively, in cells expressing rHsp90 (Fig. 7 F ). In the case of rHsp70 expression, all the viral isolates tested, except WWM, showed significantly increased percentages of infection as compared to those showed by control cells (Fig. 7 F ). Treatment of cells with the inhibitor for Hsp90 (17AAG) reduced the percentages of infection for all the rotavirus isolates tested by 40% to 71% relative to the percentages observed in control cells (Fig. 7 F ). These results confirm that Hsp90 and Hsp70 are used by the rotavirus isolates studied. Representative images for each of the treatments described in fig 7F are shown (Fig. 7 G )
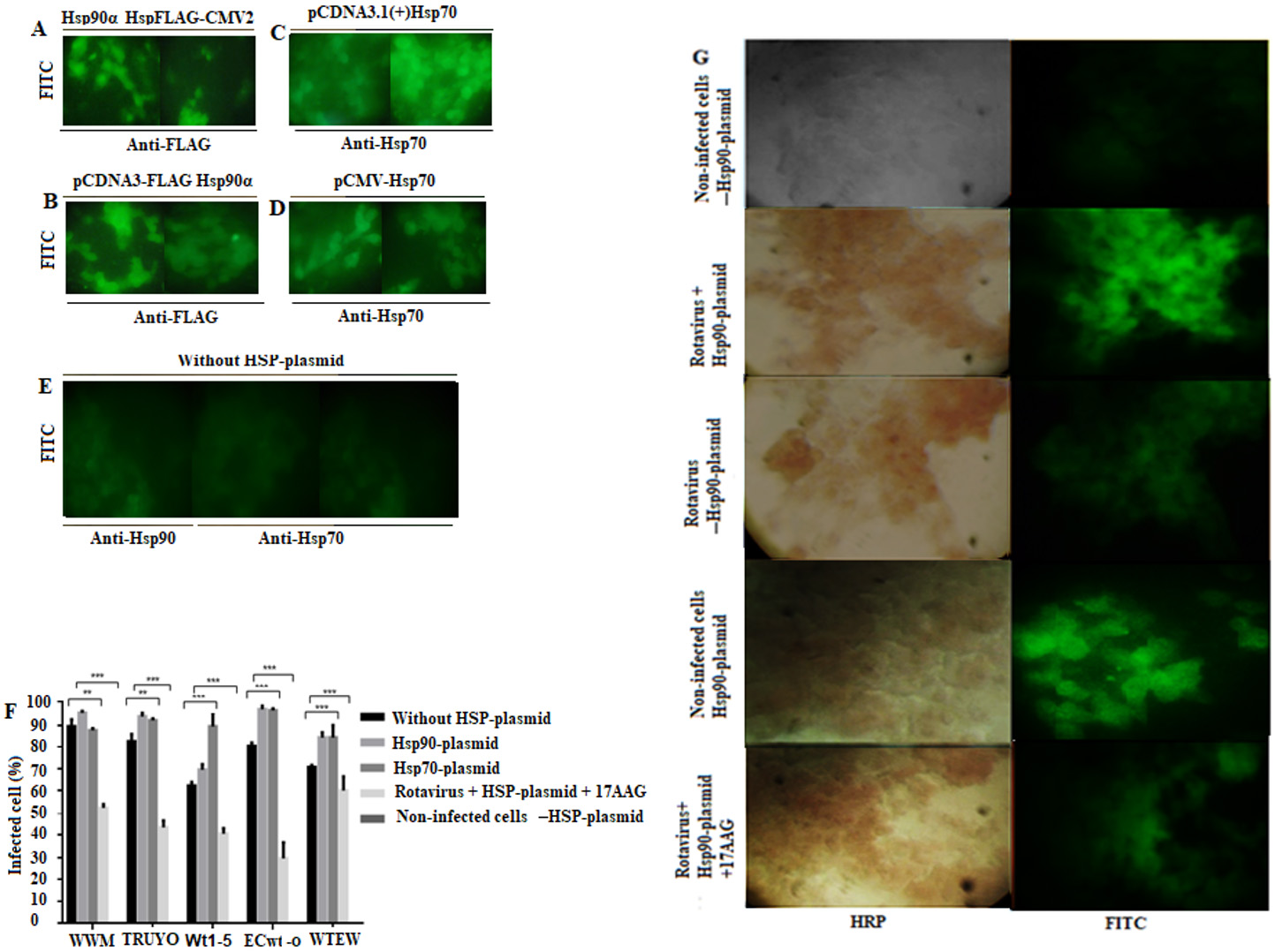
Figure 7 Increase of rotavirus infection by overexpression of rHsp90 and rHsp70. MCF-7 cells were transfected with the indicated plasmids expressing rHsp90 or rHsp70. After 24 h of transfection, the cells were infected with the indicated rotavirus isolates for 12 h at 37( C. A-E. Expression of recombinant proteins was detected by immunofluorescence using specific antibodies to them Representative photographs are shown. Non-transfected cells were used as a control. F. Transfected or non-transfected cells with Hsp90- or Hsp70-containing plasmids were separately infected or left uninfected with the indicated rotavirus isolates. Aliquots of transfected cells expressing rHsp90 were treated the inhibitor 17AAG before infection with rotavirus isolates. Cells transfected with the empty plasmid or non-transfected were used as a control. Infection was expressed as percentage of cells being positive to rotavirus structural antigens. G. Representative photographs of transfected cells infected or left uninfected with the rotavirus isolates. Rotavirus antigens were detected with secondary HRP-conjugated antibodies (reddish-brown), and recombinant HSPs (rHsp90 or rHsp70) were detected with secondary FITC-conjugated antibodies (green). 17AAG was used as an inhibitor of Hsp90. Control cells consisted of non-infected cells transfected with HSP-plasmid (either rHsp90 or rHsp70) and rotavirus infected or left uninfected cells transfected with the empty plasmid (-HSP-plasmid). Data are shown as mean percentages ( standard deviation from three independent experiments performed in duplicate.
Discussion
The last decades have been characterized by the search of alternative therapies for treating cancer since the well-known adverse effects associated with the use of traditional chemotherapy and radiotherapy. Virotherapy represents one of these alternative therapies as recent advances have shown that the use of oncolytic viruses is a safe and effective alternative 39. The use of oncolytic viruses must meet some requirements, including limited or no infection of cells other than tumor cells, which depends on their unique tropism for human tumor cells exhibiting cellular receptors that facilitate viral entry.
The present study provides evidence that a relatively high percentage of MCF-7 cells express on their cell surface the HSPs examined. The presence of HSPs in cell membrane-enriched fractions is also supported by their direct or indirect interaction with Hsc70 and integrin β3 which have previously reported as cell surface membrane protein markers 40,41. Moreover, the results obtained from the membrane enriched fractions suggest that differential quantitative interactions between virus isolates and cellular proteins are occurring. However, whether or not these interactions are direct is unclear from these particular experiments as interactions mediated by other cellular proteins cannot be excluded.
Evidence is also provided that Hsp90, Hsp70, Hsp60, and Hsp40 are involved in rotavirus infection of MCF-7 cells. The inhibitory effects on viral infection induced by pre-treating cells with antibodies to these cellular proteins suggest that they are involved in the virus entry process. This conclusion is further supported by the fact that these cellular proteins are located in lipid raft microdomains forming a complex with PDI, Hsc70 and integrin β3, which have been reported to be part of the cellular machinery involved in rotavirus entry 27. The use of F(ab’)2 fragments instead of whole immunoglobulins reduces the possibility that anti-HSP antibodies are producing a steric impediment.
In addition, the negative effect produced on virus infection by the pre-treatment of cells with specific inhibitors for Hsp90 and Hsp70 is also supporting the implication of these cellular proteins in the rotavirus infection process. In this line of thought, the increased viral infection caused by overexpression of recombinant Hsp90 and Hsp70 is also in favor of the participation of these cellular proteins in the infection produced by the rotavirus isolates tested. The identification of viral cell receptors is not only useful for designing anti-viral strategies but also for understanding viral tropism in the context of the specificity required by oncolytic virotherapy.
We used the human mammary gland adenocarcinoma cell line MCF-7 as a model system to identify some of the cell surface proteins involved in rotavirus binding that mediate viral entry. We found that the binding of rotavirus particles to the cell surface HSPs functioning as receptors triggered the viral infection which was detected in terms of the percentage of cells being positive to viral antigens or the total amount of viral antigens accumulated at a post-infection time that is compatible with at least one viral life cycle. The use of recombinant Hsp90 and Hsp70 for pre-treating rotavirus virions before inoculation of MCF-7 cells allowed us to conclude that these cellular proteins compete with their counterpart located on the cell surface for binding to rotavirus particles, leading in this way to a decreased infection. The interaction between virions and Hsp90 and Hsp70 was further evidenced by capture ELISA and co-immunoprecipitation assay.
From the point of view of receptor usage, it is very interesting to highlight that rotaviruses appear to use alternative entry pathways. In the present work, the HSPs inhibitors and antibodies to HSPs were not only unable to completely inhibit rotavirus infection, but the rotaviruses isolates tested responded differentially to these inhibitors and antibodies. It deserves to be highlighted that quercetin inhibited rotavirus infection at higher temperature. It has been reported that quercetin increases its inhibitory effect on Hsp70 at higher temperatures 42. Although the effect of inhibiting Hsc70, PDI and integrin β3 on the rotavirus infection was no tested in the present study, previous studies have shown that these cellular proteins are used for rotavirus entry into MA104 cells 40,41. However, the presence of theses cellular proteins on the cell surface of MCF-7 cells forming complexes with HSPs in lipid rafts and their probable direct interaction with rotavirus particles suggest that they might be used by the rotavirus isolates assayed in the present work. These results suggest that rotavirus seem to have evolved to use different types of cell surface molecules to enter the target cell. Chaperoning molecules, such as HSPs, in addition to binding molecules (e.g. acetylsalicylic acid, integrins) and redox molecules (e.g. PDI, Erp57) have been proposed as candidate receptors supporting alternative entry pathways 31. The presence of several cell surface proteins playing the role of receptors or co-receptors for rotavirus entry allows us to give further support to the proposed sequential and alternative interaction model 43, which includes the binding and conformational changes of rotavirus structural proteins before penetration into the target cell. However, our results do not exclude that other cell surface molecules may be helping to facilitate rotavirus entry into the host cell.
Our findings highlight that the rotavirus isolates studied use Hsp90, Hsp70, Hsp60 and Hsp40 as receptors for infecting MCF-7 cells. HSPs are a family of cellular proteins involved in chaperone activity dependent on ATP hydrolysis. Hsc70 has been reported to induce conformational changes in rotavirus particles 26. We conclude that the tumor cell-adapted rotavirus isolates studied in the present work can successfully infect the human mammary gland adenocarcinoma cell line MCF-7 by binding to Hsp90, Hsp70, Hsp60 and Hsp40 present on its cell surface. This receptor-dependent tropism exhibited by these rotavirus isolates deserves to be highlighted since receptors determine the entry of the virus into the host cell and HSPs are overexpressed in a range of human tumors 22. However, it remains to be examined whether non-tumor cells with increased heat shock response are a target of the rotavirus isolates studied here. Taken together, our findings suggest that the rotavirus isolates studied in the present work deserve to be further investigated as a potential oncolytic agent. Since essentially all population has antibodies to rotavirus, a balance between the immune anti-viral response and immune anti-tumor response should be sought.











 text in
text in 




