INTRODUCTION
Citrus fruits are recognized for their high water and vitamin C contents, along with sugars, acids, mineral salts, fibers, and other vitamins that benefit human health (Valera, 2018). They are also known as an important source of antioxidants (Zou et al., 2015) because of their high content of polyphenols, flavonoids, and terpenes. Nowadays, 140 countries harvest citrus fruits. In 2017, global production was estimated at 120 million tons, with the orange ranked as first. The mandarin had an average of 23.3 million tons (Valera, 2018). The main citrus producing countries are China, Brazil, India, and the United States, with citrus species that include the lemon, mandarin, orange, grapefruit, and lime (de la Rosa-Hernández et al., 2016).
In the citrus group, the mandarin is the second most important citrus fruit, with the highest climatic adaptation (Mora-Vivas et al., 2013). For example, China, with its varied geography, produces an average of 10 million tons per year. In Latin American, mandarin producing countries include Colombia, with an average of 110,000 t, followed by Peru, Bolivia and Uruguay (Cámara de Comercio, 2019). The hilly plains have arrayana mandarin (Citrus reticulata Blanco) crops of the Dancy variety, whose fruits have outstanding quality and production, confirming adaptation to this area (Mateus-Cagua et al., 2015).
In Colombia, citrus fruits represent 15% of the production of perennial crops, with a total planted area of 65,000 ha (Ramírez et al., 2014). It is estimated that there may be between 10 to 12 thousand hectares of mandarin in Colombia (Orduz et al., 2012). The quality of citrus fruits is very important for commercialization, including the internal and external characteristics of fruits, appearance and organoleptic quality, which are considered for consumption and industrial processes.
Mandarins grown in the Norte de Santander Department, especially in the Villa Sucre area (Arboledas municipality), are known as the "Criolla mandarin" ramet but this classification, despite its marketing aspect, is very unspecific and is not assertive. However, because of the increase in demand for fruits in the country, small farmers in this area want to increase and improve their crops to gain greater competitiveness in the market over other producers in the region and further commercialization of their products.
Tangerines are highly polyembryonic, and clonal propagation of seeds confers conservation advantages, preserving the genetic identity of the parental characteristics. On the other hand, citrus fruits are vulnerable to microorganism infections, causing physiological and physical pathologies and lowering high-quality harvest every year. As a result, chemical fungicides are widely used to control plant diseases and pests but they have side effects on human health and the environment (Palou et al., 2015). The conservation and use of plant genetic resources are essential for the improvement of agricultural production, sustainable development, and poverty management. Molecular biology analyses of genetic diversity have contributed substantially to the understanding of plant genetics (Monfared et al., 2018).
Generally, genetic diversity can be characterized using morphological, biochemical, and molecular factors (Kremer et al., 2015). Morphological and biochemical factors are affected by environmental conditions, hiding the true representation of genetic diversity, and are prone to underestimations. On the other hand, molecular DNA methods include all factors, environmental factors and the genetic portrait, displaying a more objective representation of diversity (Liu et al., 2013).
To achieve these goals, ISSRs (inter repeated simple sequences), a type of co-dominant molecular markers frequently found in most eukaryotic genomes, are used (Mei et al., 2017). Some advantages are high reproducibility, efficiency, high level of polymorphism and high information capacity (Venkat et al., 2014), making them suitable for differentiation and identification of closely related genotypes (Cui et al., 2017; Souza et al., 2017). Therefore, they have been widely used in genetic diversity characterizations (Heidari et al., 2016; Tiwari et al., 2016), ramet identification (Zheng et al., 2011), taxonomy (Zvyagina et al., 2016), phylogeny and molecular mapping (Tomar et al., 2017). They are also useful for detecting intraspecific genetic variations (Poyraz, 2016).
Therefore, given the need to improve mandarin (Citrus reticulata) business for farmers and the impacts on local and global economies in Colombia, this study characterized the genetic diversity and genetic relationships among ten populations of C. reticulata distributed mainly in the village of Villa Sucre using ISSR markers. The main objectives were to establish the first molecular characterization of mandarin (C. reticulata) and to assess the genetic variation and differentiation within and between populations.
MATERIALS AND METHODS
Ramet selection and DNA samples
Ten mandarin ramets (C. reticulata) were selected from ASOMUSUCRE farms, a union that oversees the Villa Sucre area. All commercial ramets older than 5 years and geographically close farms were considered. The sampling on each farm was as follows: three trees were chosen randomly, and healthy leaves were sampled and stored in wet towels and then in plastic bags at -20°C. Ramet samples were also collected from a commercial farm, located in the Arboledas area (Eden Farm), where a representative sample was collected (N = 31) (Fig. 1).
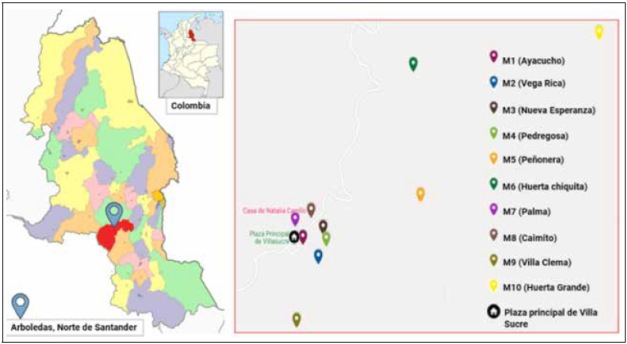
Figure 1 C. reticulata farms in Villa sucre (Arboledas’ Municipality, Norte de Santander, Colombia). Sampling locations are indicated by colored drop mark. Six citrus farms were located around Villa Sucre’s downtown central square, and the other five were located in the town’s rural area. Farm sampling codes, Ramet age and plantation type: M1 = Ayacucho (18 years/graft); M2 = Vega Rica (5 years/seed); M3 = Nueva Esperanza (25 years/graft); M4 = Pedregosa (20 years/graft); M5 = La Peñonera (12 years/graft); M6 = Huerta Chiquita (15 years/graft); M7 = La Palma (15 years/graft); M8 = El Caimito (15 years/graft); M9 =Villa Clema (5 years/seed); M10 = Huerta Grande (15 years/graft); Mx = Eden (40 years/graft - Commercial Ramet).
DNA extraction
The genetic material isolation was carried for every sampled tree: five young leaves were ground in liquid nitrogen, and DNA extraction was done following the Suárez and Osorio (2017) extraction protocol. The extracted DNA was then quantified using a NanoDrop 2000 (Thermo Scientific™), and the integrity was assessed with 0.8% agarose gel electrophoresis and visualized with a ChemiDoc™ MP Imaging System (Bio-Rad). Finally, the samples were diluted to a final concentration (25 ng μL-1) and stored at - 4°C.
ISSR markers amplification and visualization
The DNA amplification was done with a 25 µL final PCR reaction volume for each primer (12.5 μL og GoTaq® Green Master Mix (Promega CO) 0.5 μL primer, 10 μL nuclease-free water, and 2 μL DNA). The final primer concentration for amplification was 0.25 µM, as suggested by Medina-Medrano et al. (2016) and Latorre et al. (2013). The PCR was done in a MultiGene™ OptiMax (Labnet) thermocycler, and the thermal conditions were: initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 60 s, 49 - 64°C for 120 s depending on the ISSR primer (Tab. 1), 120 s at 72°C and a final extension at 72°C for 7 min. DNA Amplicons were confirmed in a high resolution 2% agarose gel at 80V for 2 h, stained with SYBR Green and later visualized in a ChemiDoc™ MP Imaging System (Bio-Rad Laboratories, Hercules, USA). The gel-band pattern was used to assign the genotype profile for every ramet.
Table 1 ISSR primers used for the analysis of C. reticulata (Medina-Medrano et al 2016;Latorre et al., 2013).
| Primer | Primer sequence | Standardized Tm |
|---|---|---|
| 827 | (CA)7GC | 49.8 |
| 864 | 5’-ATG ATG ATG ATGATG ATG-3’ | 46.5 |
| 878 | GGA TGG ATG GAT GGA | 46.5 |
| 880 | 5’-GGA GAG GAG AGG AGA-3’ | 50.0 |
| 888 | 5’-CGTAGTCGTCAC ACA CAC ACA CA-3’ | 64.7 |
| 890 | 5’-ACG ACT ACG GTG TGT GTG TGT GT-3’ | 64.7 |
Data analysis
Electrophoretic bands were considered single molecular characteristics, used to construct a binary matrix encoded as 1 (presence) or 0 (absence) for each farm population sample. The binary data were later used to calculate the percentage of amplified polymorphism, dividing the number of polymorphic bands by each primer sample’s total bands. The informative and discriminatory capacity of each primer was evaluated with the proportion of polymorphic information content (PIC) and calculated with the equation: PIC = 2 Pi (1 - Pi), where, Pi is the occurrence frequency of the polymorphic band for each primer (Roldán-Ruiz et al., 2000). The Dice similarity index was determined using InfoGEN (2013) (Balzarini and Di Rienzo, 2013). The relationship the between ramets was evaluated by constructing a dendrogram using the UPGMA algorithm and a similarity matrix based on the Dice correlation cophenetic coefficient (CCC). This coefficient was assessed between different similarity indices (Jaccard, Euclidean 2 and Dice) at its highest value (r = 0.89) to test the best good fit. Binary information was used to display the genetic variation in a multidimensional scaling Plot (MDS). AMOVA analysis was carried out to evaluate the molecular variation within the ramets.
RESULTS AND DISCUSSION
ISSR amplification
The ISSR primers generated a total of 61 amplified fragments, with an average of 10 amplicons per primer for the 31 samples (Tab. 2). The amplified product sizes ranged between 300 and 3.500 bp, as reported for other Citrus species with ISSR (Kumar et al., 2010).
Table 2 Number of amplified bands, number of polymorphic bands, percentage of polymorphism, PIC value and amplification range of each primer.
| Primer | No. of bands | % polymorphism | PIC | Amplification range |
|---|---|---|---|---|
| 827 | 8 | 39 | 0.47 | 400 - 1,300 Pb |
| 864 | 9 | 44 | 0.49 | 400 - 1,500 Pb |
| 878 | 14 | 52 | 0.5 | 600 - 3,500 Pb |
| 880 | 6 | 75 | 0.38 | 1,200 - 2,500 Pb |
| 888 | 11 | 20 | 0.32 | 300 - 2,500 Pb |
| 890 | 13 | 23 | 0.35 | 300 - 2,500 Pb |
| Mean | 10 | 42 | 0.42 |
The average PIC value observed for the six primers was 0.42. For dominant markers such as ISSR, the maximum value is 0.5 (Sehgal et al., 2009). In the present study, every primer recorded a representative genetic variability across the samples; the 878 primer displayed the highest PIC value (0.5), indicating the usefulness of this marker for variation characterization in C. reticulata. Primer 888 (Fig. 2) revealed the lowest polymorphic information content (0.32). Primers 880 and 864 (Fig. 3) were the most useful for identifying two samples that shared the same allele, indicating that these primers had a high level of confidence for discriminating between the analyzed C. reticulata species.
Genetic diversity analysis
Among the several indexes of similarity, the Dice coefficient was chosen as the most appropriate for the ISSR markers since it has no genetic meaning with the coincidence of missing bands (Alburquerque et al., 2008). The range of the Dice similarity coefficients varied from 0.0 to 0.68. The lowest genetic similarity was between M1b/M2c and M1c/M2c, with a coefficient of similarity of 0.68. M1b/M1c and M7a/M7b were very similar genetically, with a 0.0 coefficient. The genetic differentiation value indicated a more significant proportion of the genetic variation within the populations, as indicated by the AMOVA analysis (89.6%). According to the Dice similarity matrix, the samples obtained a high genetic correlation (0.46) on average. These results were probably due to the fact that the samples of each species were taken in triplicate for each crop and were collected in the same geographical area (Villa Sucre), meaning a large part of the C. reticulata samples are genetically related and there was little genetic diversity within the studied populations. These results suggest that, although the C. reticulata samples were geographically and genetically related, there was moderate genetic diversity within the populations.
Relationship between populations
The correlation coefficient (r = 0.89) indicated good fit between the clusters in the dendrogram and the matrices of similarity. The dendrogram grouped the 31 C. reticulata samples into five groups and two subgroups (Fig. 4). Group 1 had the highest number of samples in 2 subgroups, where the first subgroup (Ia) contained the standard sample (Mx) and the Nueva Esperanza farm (M3), indicating that this ramet displayed the highest genetic variation, followed by the commercial criolla mandarin ramet. The second (Ib) subgroup held the M4 and M5 samples, from the Pedregosa and La Peñonera farms. Group 2 brought together samples M7 (la Palma) and M8 (el Caimito). Samples M2 (Vega Rica) and M9 (Villa Clema) formed independent groups (Group V and III). The ramets from these farms were different from the others (plantation type), developed by sexual reproduction (seeds). M6 (farm H. Chiquita) and M10 (farm H. Grande) did not group with other samples, with samples M6c and M10a included in Group IV. In general, the molecular results confirmed the classifications of the samples based on the sampling zones and the morphological characterization.
For instance, most of the samples from the Villa Sucre farms tended to associate in a single group (M1, M2, M3, M4, M5, M7, M8, M9). The analyses showed little genetic variation within the species but moderate variation between them. Hogbin and Peakall (1999) suggested that a low level of population differentiation can be explained by several factors, including the reproduction system of the species, genetic drift, or the geographical isolation of the populations. The analyzed C. reticulata ramets are frequently gregarious in nature and not only proliferate through seeds but also through shoots. Therefore, populations in nature are more homogeneous.
The MDS analysis calculated with the amplification profiles of the 6 ISSR primers (Fig. 5) explained the variation in two dimensions. These dimensions explain 47.5% of the total variability and revealed five groups, in which individuals had a clear association pattern, allowing specific discrimination. The coordinate analysis supported the UPGMA dendrogram cluster pattern, revealing that the spatially close populations tended to be genetically similar (M7/M8) and the more distant populations had little genetic similarity (M1/M2). Since the species had crossbreeding and most of the C. reticulata populations were less than 1 km away, the geographical distance between the samples may have played an important role in the differentiation between the populations. Sarwat and Srivastava (2008) indicated that geographically isolated populations accumulate genetic differences as they adapt to different environmental conditions. Likewise, Zong et al. (2008) suggested that, in small and isolated populations, genetic drift influences the genetic structure and increases the differentiation between populations.
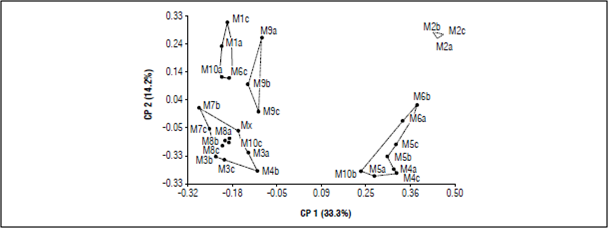
Figure 5 Multidimensional scaling plot analysis for 31 samples of C. reticulata in the Villa Sucre area.
Although there are few studies in the Norte de Santander region of Colombia there are several genetic diversity studies from the Boyaca Department that used RAM (Random Amplified Microsatellites) markers on Solanum quitoense Lam. The data showed high genetic variability for the area and suggested the need for subdivision levels in the sampled ramets that would allow better use of the genetic diversity in S. quitoense from Neira’s province. Moreover, Morillo-Coronado et al. (2017) used seven RAMs to show that the cape gooseberry genetic diversity (Physalis peruviana L.), an intraspecific variation that must be conserved, for use in genetic improvement programs for adapted materials with high quality agronomic, nutritional and medicinal characteristics (Morillo-Coronado et al., 2018). In addition, 30 mandarin (Citrus reticulata) accessions from the Corpoica-Palmira germplasm bank have been molecularly characterized with RAMS, random microsatellites, a technique that groups mandarin accessions according to the reported types (Mandarino Satsuma, Mandarinos Clementinos and Hybridos) and is a useful molecular tool for assessing genetic diversity in Citrus (Mora-Vivas et al., 2013).
According to Lezcano (2018), SSR markers are more effective than RAPD in closely related Citrus varieties. This author suggested these markers are useful for elucidating genetic diversity, determining kinship, and revealing phylogenetic relationships between various citrus species.
ISSR applications (Inter-Simple Sequence Repeats) revealed polymorphisms in other studies on oranges that made it possible to unequivocally differentiate varieties of navel oranges and grapefruits. The RAM (Random Amplified Microsatellites) technique grouped accessions of white oranges, navel oranges and grapefruits. The results can be used to identify varieties of navel oranges. Molecular markers that allow for the unequivocal distinction of varieties of white oranges and grapefruits have not been found (Fernández, 2018).
Other studies on citrus in other latitudes with ISSR molecular markers have shown an average percentage of polymorphic bands of 42%, which is comparable with other results for citrus. For example, 15 ISSR primers revealed a polymorphism of 54, 55 and 56% in the species Citrus indica tanaka, Citrus latipes, and Citrus medica, respectively (Kumar et al., 2010).
Joshi and Dhawan (2007) observed 42.5% polymorphism among 13 genotypes of Swertia chirayita. However, other authors have observed higher percentages of genetic diversity in terms of the polymorphic information content, ranging from 85 to 100% (Alburquerque et al., 2008; Marak and Laskar, 2010; Rahimmalek et al., 2009; Shahsavar et al., 2007; Shilpha et al., 2013).
CONCLUSION
The present study estimated the genetic variation between populations of C. reticulata from the Villa Sucre area (Norte de Santander). The ISSR primers were useful for differentiating closely related species, demonstrating the lack of classification and identification studies on citrus fruits in Norte de Santander and in Colombia.
The results also showed that the analyzed samples were intermingled, forming different groups independent of their geographical affiliations. This pattern of distribution of genetic variation among the samples could have been generated by the free exchange of germplasm between the samples and the reproduction method used on the farms. On the other hand, the small intraspecific genetic variation could be explained by the fat that citrus species hybridize easily.
C. reticulata has been widely used in citrus marketing systems in Colombia for a long time. Despite its high potential, little or no attention has been paid so far to preserving this species. Therefore, the present study determined the level of genetic variation between C. reticulata populations in the Villa Sucre district (Norte de Santander). The ISSR primers were an effective tool for differentiating closely related species, confronting the problem of citrus classification and identification in Norte de Santander and in Colombia.





![Validation of black spot [(Pseudocercospora purpurea (Cooke) Deighton] management strategies in avocado crops in northern Tolima (Colombia)](/img/en/next.gif)












