Introduction
Cyanobacteria are a group of photosynthetic prokaryotes that are among the oldest and most successful forms of life, with a diverse range of functional traits that allow them to colonize various environments and cope with adverse conditions (Dalu and Wasserman, 2018; Roque et al., 2021). In tropical regions, permanently high temperatures in aquatic ecosystems have been demonstrated to promote the dominance of this phylum of phytoplankton over other forms throughout the year (Chorus and Bartram, 1999; Dalu and Wasserman, 2018; Harke et al., 2016; Paerl et al., 2011; Reynolds and Petersen, 2000).
Several studies suggest that surface sediments act as reservoirs for cyanobacteria in aquatic ecosystems and indicate that the benthic zone may be an important source of cyanotoxins, even in the absence of planktonic surface cyanobacteria (Bormans et al., 2020; Gangi et al., 2020; Izaguirre et al., 2007; Westrick and Szlag, 2018). Furthermore, benthic environmental samples collected in warmer regions have been found to contain significantly more toxins than those collected in temperate climates (Gaget et al., 2022), underscoring the necessity of advancing our understanding of their ecology to comprehend their dynamics in tropical freshwater systems (Quiblier et al., 2013).
Most investigations on benthic cyanobacteria in tropical and subtropical regions have been conducted in shallow environments (Borges et al., 2015; Crispino and Sant'anna, 2006; Magina and da Silva e Silva, 2008; Noffke et al., 2003; Silva-e-Silva, 2004), and the knowledge of these organisms in high mountain reservoirs is still limited. The overall perspective suggests that research on benthic cyanobacteria is lagging, and the risk associated with them remains largely unknown (Gaget et al., 2022); Colombian reservoirs are not far from this problem.
Although the increase in eutrophication and water temperature favors the proliferation of cyanobacteria in tropical aquatic ecosystems, there is no systematic record of cyanobacterial blooms or a protocol for their control and management in Colombia (Salomón et al., 2020). Current knowledge about the taxonomy and ecology of cyanobacteria in Colombian reservoirs is mainly limited to taxonomic lists of planktonic species, while the ecological characteristics and toxic potential of cyanobacteria from the benthic zone are unknown.
These knowledge gaps were highlighted by Quiblier et al. (2013) and more recently by Gaget et al. (2022) in their respective studies, each proposing that one reason for this is that water utilities rarely establish monitoring practices for benthic cyanobacteria or associated secondary metabolites.
The present study aimed to elucidate the role of surface sediments as a reservoir of cyanobacterial with toxic potential in two tropical reservoirs. For this, we conducted an initial germination experiment with sediments samples and evaluated their viability and toxic potential, as described in Arismendi-González et al. (2021). Subsequently, we performed molecular analyses on the cultured samples to gain insights into the richness, diversity, and composition of the cyanobacterial community in the surface sediments of both reservoirs.
These analyses were carried out under laboratory conditions, and our main objective was to establish a baseline for future research on the ecology and potential risks associated with benthic cyanobacteria in these systems. Our findings provide valuable information on the presence and characteristics of cyanobacteria in two tropical reservoirs and may aid in the development of strategies for their management and control.
MATERIALS AND METHODS
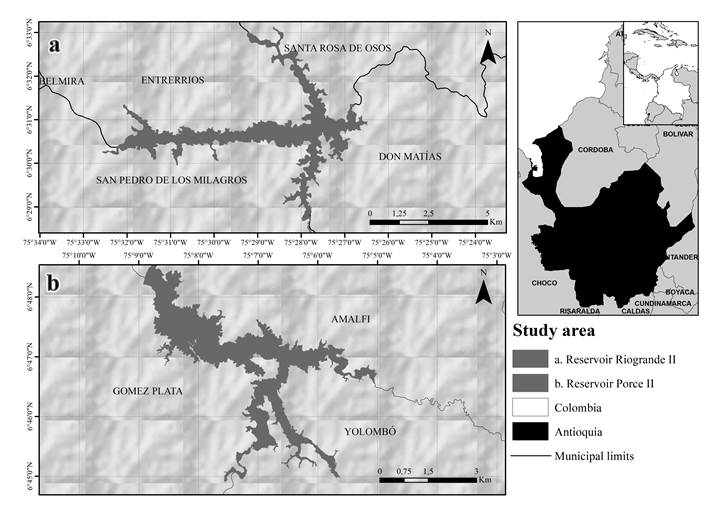
Study area
Riogrande II reservoir is located northwest of Medellín (Antioquia), Colombia (75°32'30'', 75°26'10''W and 6°33'50'', 6°28'07'' N) with a maximum flood level of 2270 m.a.s.l. It is mainly used for drinking water withdrawal, but also for electric power generation (up to 321MW). This system has a surface area of 12.14 km2, with a total volume of 240 million m3, and a maximum depth of 47.2 m. The water temperature span ranges from 15 °C to 25 °C and the mean annual rainfall of the area is approximately 2250 mm (Figure 1a).
Porce II reservoir is located northwest of Antioquia, Colombia (75°09'14", 75°04'59" W and 6°44'57", 6°48'45"N) with a maximum flood level of 925 m.a.s.l. It is mainly used for electric power generation (405MW). The reservoir has a surface area of 9.8 km2, with a mean volume of 231 million m3, a maximum depth of 96.5 m and the mean annual rainfall of the area is approximately 1960 mm (Figure 1b).
Sampling
From December 2014 to September 2015, sediment samples were collected at three stations in each reservoir (Figure 1). A UWITEC gravity nucleator equipped with transparent acrylic tubes, approximately 50 cm in length and 6 cm in diameter, was employed for sediment sampling collection. All samples were kept at 4 ºC. Triplicate samples were obtained from each station of the same reservoir at all four sampling events.
Laboratory analysis
Cyanobacterial cultures from sediment samples
Cyanobacterial cultures were established from environmental samples collected from the oxic and silty upper layer sediments (0-4 cm). Five g of sediment samples were aseptically transferred into sterile 250 mL glass bottles containing 50 mL of ASM-1 medium. Cultures were incubated at 25 °C in a climate chamber using dark/light (1300 lx) cycles of 12 h, as previously described by Arismendi-González et al. (2021). The cultures were monitored every 20 days. However, the evaluation of the cultures was conducted at the end of the experiment, 300 days later.
Identification of cultured cyanobacteria from sediment samples
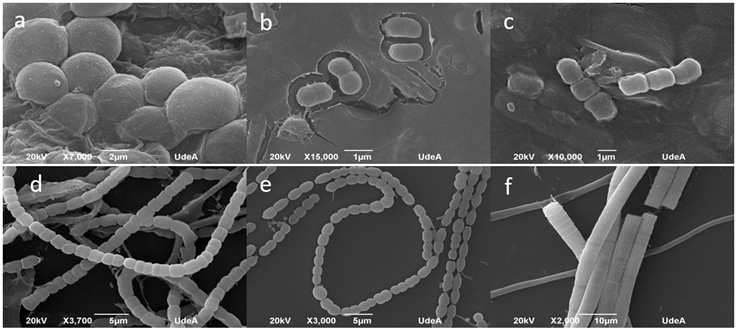
To confirm the presence and viability of cyanobacteria in the cultures, chlorophyll-a measurements were performed using the FluoroProbe III Workstation 25 (bbe-Moldaenke, Kiel, Germany) following the methodology outlined in Arismendi-González et al. (2021). This allowed us to differentiate each phytoplankton group and verify cyanobacteria growth in each culture sample. To identify cyanobacteria, we used an Olympus CX31 microscope at 400X magnification. Additionally, random samples from the successfully isolated cultures were analyzed using a JSM-6490LV Scanning Electron Microscope-SEM (Jeol, USA). Identification was performed following the Komárek and Anagnostidis (1998; 2005), Komárek (2013), and Tucci et al. (2019) taxonomic keys.
Quantification of Microcystins by ELISA in culture from sediment samples
Microcystins (MCs) production by cyanobacterial cultures was assessed by enzyme-linked immunosorbent assay (ELISA) with the Envirologix QuantiPlate commercial kit (Envirologix Inc., Portland, ME, USA). The ELISA assay had a limit of detection between 0.16 to 2.5 ppb. Before analysis, all samples were subjected to three freeze/thaw cycles to disrupt the cellular matrix and promote toxin release. The samples were further processed by sonication using a Branson model 2510R-DTH Ultrasonic Cleaner (Branson Ultrasonic Corp., USA) to facilitate cell lysis and toxin release, then ELISA test was performed following the manufacturer's instructions.
Molecular analysis of cyanobacterial diversity in cultures from sediment samples
Sub-samples from each reservoir (viable cultures from all sampling stations were included) were pooled and prepared for DNA extraction. Total genomic DNA was extracted from each sub-sample using a genomic DNA mini kit (Invitrogen™, K1820-02) according to manufacturers' recommendations, and DNA concentration was assessed using Quant-it PicoGreen dsDNA Assay (Invitrogen™, USA).
Amplification and sequencing of 16S rRNA gene by Next Generation Sequencing-NGS
The 16S rRNA gene was full-length amplified (1500pb) using the primers 27F1 (5’ AGAGTTTGATCCTGGCTCAG 3’) y 1492R (5’ TACGGYTACCTTGTTACGACTT 3’) (Heuer et al., 1997) and the quality of the amplicons was assessed by 1% agarose gel electrophoresis. The samples were sequenced by the Illumina MiSeq platform generating paired reads of 300pb. The V3-V4 hypervariable regions from the 16S rRNA gene were amplified by PCR using the universal bacterial primers Bakt_341F (5’ CCTACGGGNGGCWGCAG 3’) and Bakt_805R (5’ GACTACHVGGGTATCTAATCC 3’) (Herlemann et al., 2011), both with Illumina adapter, pad, and linker sequence.
16S amplicon sequence analyses
Sequences were analyzed using the Mothur software (Schloss et al., 2009) according to Miseq Standard Operating Procedure. The quality control analysis of the sequencing data was performed through the FastQC program, selecting only the sequences with a Q value > 30 and the VSEARCH program was applied to identify and remove the chimeric sequences. The Operational Taxonomic Units (OTUs) were defined with a minimum of 97% identity by taxonomy assignment using the RDP reference database sequences as a guide. In addition, the OTUs representative sequences were analyzed using BLASTn on the NCBI database. The candidate cyanobacterial sequences were selected with a minimum 95% identity in the BLASTn using a 16S cyanobacterial database. Through Mothur software, richness (Chao index) and diversity (Shannon index) were calculated by using the number of observed OTUs, and sample coverage was calculated.
Phylogenetic analysis of culturable cyanobacterial strains
To compare cyanobacteria species from the Riogrande II and Porce II reservoirs a phylogenetic analysis was constructed using the MEGA 7.0.21 software. The nucleotide sequences used have at least 95% identity to Operational Taxonomic Units (OTUs) identified through 16S amplicon sequence analyses conducted on sub-samples of cultures from surface sediment of each reservoir. The Maximum likelihood method was performed to build the phylogenetic tree, using Kimura 2-parameters model plus gamma distribution with invariant sites. To estimate the confidence of the branching patterns of the tree, a bootstrap test was executed with 1000 replications. In total, 112 sequences were analyzed, comprising 51 sequences obtained in this study along with 61 representative sequences from the most abundant groups, as well as the closest relatives determined through BLASTn. All nucleotide sequences from this study were deposited in the GenBank database under accession numbers OR452809 to OR452855.
Results
Cyanobacterial cultures from sediment samples
Spectral analysis revealed the presence of cyanobacteria in the surface sediments of both reservoirs, with mean concentrations of chlorophyll-a cyano of 591 μg/L and 1172 μg/L respectively. Furthermore, microscopic analysis of the cultures enabled the identification of different genera, corroborating their viability and providing insights into the composition and diversity of the cyanobacterial communities in each reservoir.
The composition of cyanobacterial communities in Porce II and Riogrande II reservoirs are shown in Table 1. The microscopic assessment revealed the presence of four orders (Figure 2) and a total of 27 genera, with 16 of them identified in cultures from both reservoirs. The genera Merismopedia, Fischerella, and Hapalosiphon were identified exclusively in Porce II, while Limnothrix, Pseudanabaena mucicola, Pseudanabaena galeata, Aphanocapsa incerta, Asterocapsa, Gloeothece, Nostoc, and Anabaena were observed only in Riogrande II.
Table 1 Cyanobacterial identification by Optical Microscopy from sediment cultures of Porce II and Riogrande II reservoirs
| Orden | Nostocales | Synechococcales | Chroococales | Oscillatoriales | |||||||||||||||||||||||
| Taxa | Dolichospermum sp. | Cylindrospermum cf. | Sphaerospermopsis cf. | Nostoc sp. | Chlorogloeopsis fritschii | Calothrix sp. | Fischerella sp. | Anabaena sp. | Hapalosiphon cf.intricatus | Leptolyngbya sp. | Limnothrix sp. | Pseudanabaena mucicola | Pseudanabaena moniliformis | Pseudanabaena galeata | Aphanocapsa annulata | Aphanocapsa delicatisima | Aphanocapsa incerta | Merismopedia | Aphanotece sp. | Microcystis sp. | Chroococcus sp. | Asterocapsa | Gloeothece | Phormidium sp. | Planktothrix sp. | Geitlerinema splendidum cf | Oscillatoria sp. |
| PII | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | ||||||||
| RGII | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | x | |||
Quantification of Microcystins by ELISA in culture from sediment samples
Microcystins were detected by ELISA assay in the cultures of both reservoirs. However, in the Riogrande II, microcystin was only detected in cultures from one of three sampling stations with values close to the detection limit (0.23 and 0.27 μg/L), while in Porce II microcystin was detected in cultures from the three stations, with concentrations of MCs that ranged between 0.16 and 2.5 μg/L (Table S1).
Genetic diversity of cyanobacteria in culture from sediment samples
Community analyses of OTUs sequences
Bioinformatic analysis using the Mothur package yielded 34,530 reads in Porce II and 20,479 reads in Riogrande II, with coverage value of 0.961 and 0.963, respectively (Table 2).
Table 2 Microbial community diversity indices in sediment cultures from Porce II and Riogrande II
| Samples | Number of Reads | Coverage | Number of OTUs (richness) | Simpson index | Shannon index | Chao index |
|---|---|---|---|---|---|---|
| PII | 34,530 | 0,961 | 2183 | 24,715 | 4,718 | 6328,114 |
| RGII | 20,479 | 0,963 | 1446 | 46,746 | 5,119 | 3029,757 |
The Shannon index from both reservoirs’ samples showed that Riogrande II exhibited a higher diversity compared to Porce II. Furthermore, Riogrande II demonstrated a greater uniformity of species, as indicated by a Shannon evenness value of 0.70. In contrast, the Chao index revealed that Porce II had the highest richness of microbial communities among the two reservoirs.
The dominant phyla in the upper sediment microbial community of Porce II and Riogrande II reservoirs were Cyanobacteria, Proteobacteria, and Planctomycetes, which accounted for almost three-quarters of all reads. It should be noted that the relative abundance of Cyanobacteria was high in both reservoirs, compared to the total microbial community (Figure 3).
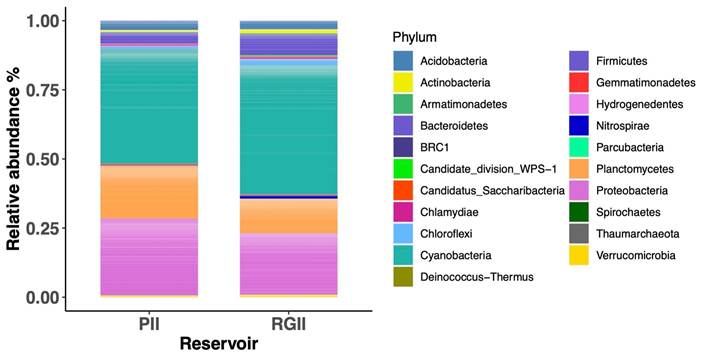
Figure 3 Relative abundance of different prokaryotic phyla (Bacteria and Archaea) in sediment cultures from Porce II and Riogrande II reservoirs.
The diversity and richness of cyanobacteria were evaluated based on the abundance of Operational Taxonomic Units (OTUs) belonging to the Cyanobacteria phylum. Out of the total 3629 OTUs acquired, 671 were identified as cyanobacteria through the NCBI database. In addition, to uphold the integrity of our final analysis, we exclusively considered sequences that demonstrated an identity percentage of at least 95%.
As seen in Figure 4, the cyanobacterial community diversity was similar for both reservoirs. However, cyanobacterial OTU richness was higher in Porce II reservoir. Of the 342 OTUs reported for Porce II, 22 were found to be shared with Riogrande II, representing the genera Anabaena, Arthronema, Calothrix, Cyanobacterium, Desmonostoc, Leptolyngbya, Nostoc, Pantalinema, Phormidesmis, Phormidium, Pseudanabaena, Roholtiella, and Tolypothrix.
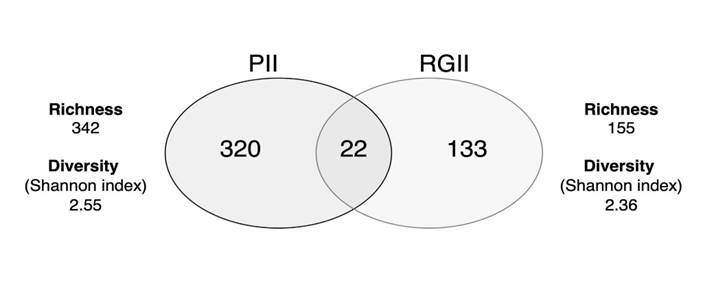
Figure 4 Venn diagram showing the number of shared and unique cyanobacterial OTUs in the Porce II and Riogrande II reservoirs, as well as their diversity and richness of cyanobacteria as a function of OTUs abundance.
According to Table S2, the highest number of OTUs in both reservoirs belonged to the orders Nostocales and Synechococcales. However, the most dominant OTUs were assigned to the genus Cyanobacterium (87) in Porce II and Leptolyngbya (42) in Riogrande II.
Phylogenetic analysis of culturable cyanobacteria strains
The phylogenetic analysis indicated that the cyanobacterial communities in these reservoirs are represented in four orders (Nostocales, Chroococcales, Oscillatoriales, and Synechococcales), as observed through the morphological analysis of the cultures. Moreover, the outcomes of this study revealed that a substantial portion of the cultivable cyanobacteria present in sediment samples from both reservoirs probably belonged to the Nostocales and Synechococcales orders (Figure 5).
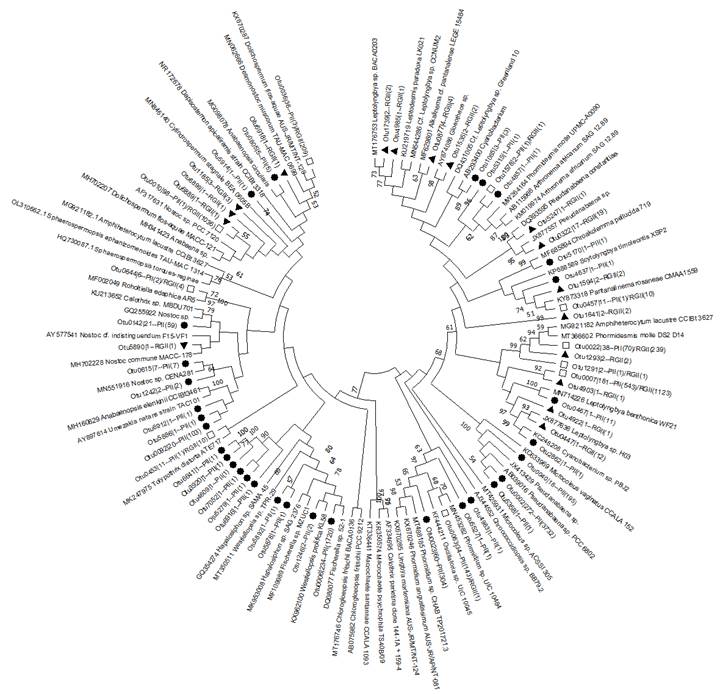
Figure 5 Phylogenetic tree based on the variable region V3 and V4 of the 16S ribosomal gene of the OTUs corresponding to the Porce II and Riogrande II samples. The shapes represent their distribution on each deposit. Circles show OTUs from Porce II, triangles show OTUs from Riogrande II, and square represents shared sequences.
Discussion
In this work, we analyzed the viability, composition, diversity, and potential toxicity of cyanobacterial species upper sediments of two tropical reservoirs: Porce II and Riogrande II. Both reservoirs are relatively deep (47 m on average for Porce II and 30 m for Riogrande II) with trophic levels between eutrophic-hypereutrophic, which reflect the high impact of human activity on these bodies of water. Previous studies have shown that the surface sediments of both reservoirs are enriched with nitrogen, phosphorus, and organic matter (Palacio et al., 2017), which leads us to think that these elements do not limit the development of phytoplankton communities in reservoirs (Arismendi-González et al., 2021).
Although it is important to consider that the study of benthic cyanobacteria is more complex, given that their growth patterns are not homogeneous within the culture containers (Quiblier et al., 2013) which could lead to erroneous estimates of the true abundance of the community, our results show how some communities of cyanobacteria settled down at the bottom of the reservoirs, survived, and were still viable, as reported by previous studies by Everson et al. (2011) and Gangi et al. (2020). Moreover, spectral analyses of the cultures showed that the mean concentration of chlorophyll-a corresponding to the cyanobacterial communities was higher in Riogrande II, which was consistent with a higher species richness observed for this reservoir through the microscopic analysis of cultures.
Despite the quantification of microcystins in cultures from upper sediment samples in both reservoirs was low, we consider that it constitutes a warning about the possible toxic potential of these populations, a hypothesis that is supported by the detection and quantification of mcy genes in direct upper sediment samples of both reservoirs as shown in Arismendi-González et al. (2021), and in other results not yet published (Table S3). Additionally, as shown in Bormans et al. (2020), the sole presence of mcy genes in sediment samples suggests that the populations present may pose a risk to organisms due to their toxic potential and their resuspension potential in the water column. However, these results lead us to consider the possibility that cyanobacterial populations present in the sediments of these reservoirs could be producing other types of toxins not included in this study, which is why we suggest a broader toxin screening be considered in future research.
According to the analysis of OTU sequences, the upper sediment samples from both reservoirs showed a high diversity of microbial communities. However, the sample from Porce II reservoir exhibited the highest species richness. On the other hand, the relative abundance of cyanobacteria was high with respect to the whole community found in culture sediment samples from both reservoirs. This finding is significant as it strengthens the hypothesis suggesting that the surface sediment constitutes an important sink for cyanobacteria, as indicated in a study conducted by Wood et al. (2021), who propose that the presence of cyanobacterial 16S rRNA genes in sediment cores provides strong evidence for cyanobacterial sedimentation in reservoirs. Furthermore, the coexistence of benthic and planktonic strains within the clades suggests that germinated cyanobacteria may belong to both habitats.
Although many of the taxa detected through phylogenetic analysis have planktonic habits, they do not totally coincide with the planktonic taxa previously described for these ecosystems (Arboleda-Baena, 2017; Palacio et al., 2015; Rodriguez, 2013; Sepúlveda-Sánchez et al., 2021). Similar results are described in the study by Wood et al. (2021) who found that none of the strains from their study were very closely related to the planktonic as well as toxin-producing taxa isolated from the Baltic Sea and various lakes in Finland, previously reported by Lyra et al. (2001) and Rajaniemi et al. (2005).
Finally, the microscopic and molecular analysis coincided on the presence of cyanobacteria of the orders Nostocales, Chroococcales, Oscillatoriales, and Synechococcales in the sediments of both reservoirs, with a greater predominance of Nostocales, which may be related to the high number of akinetes that was observed in some culture samples during germination experiments. Similar results are described in the studies of Hellweger et al. (2008) and Legrand et al. (2017a). These findings are relevant as they may indicate past blooms and contribute to knowledge about the formation of future blooms, as several studies have reported that these structures could persist and germinate from aquatic sediment samples (Gangi et al., 2020; Legrand et al., 2017b; Legrand et al., 2019; Wood et al., 2009; Wood et al., 2021) after several years. In addition, this is the first time that Fisherella and Hapalosiphon have been reported in the Porce II reservoir.
This was the first study to investigate benthic archives of cyanobacteria in the upper sediments of two Colombian reservoirs, allowing us to demonstrate through different methods the presence of active populations that reflect how these sediments provide a favorable environment for their development and possibly allow them to maintain their activity over time, which could have implications not only related to the quality of the water and the ecology of the reservoir for their possible implications in the formation of superficial blooms but also their possible repercussions on Public Health. However, this topic has not yet been explored in depth, which is why we recommend developing new research to confirm whether sediment cores can function as a proxy to predict possible blooms of planktonic cyanobacteria in reservoirs. Finally, this research highlighted the importance of further studies to achieve a holistic understanding of benthic cyanobacteria dynamics in high-mountain tropical reservoirs.