Introduction
The genome size and ploidy level varies extensively among organisms and various species. Among the flowering plants for instance, there are intra- and inter-species variation in genomic size and ploidy status among the members of a family, genus and species (Dolezel et al., 1992). This has given rise to speculations regarding the ancestral genome size of these plants and trends in their genome evolution. The genome size and genomic ploidy level, chromosome number and size variation between and within a species may be of great importance in the study of evolution and development of a breeding programme (Bennett and Leitch, 1995). In addition, the knowledge of genomic size and ploidy status of a plant could provide clues on the mechanisms responsible for decrease or increase in genomic content in the evolutionary pathway (Bennetzen and Kellogg, 1997).
Variation in chromosome number among the members of a genus of flowering plants are variously documented (Martel et al., 1997; Techio et al., 2006; Ogra et al., 2009). Also, intraspecific variations in genome size have been reported in maize (Rayburn et al., 1989) and pea (Arumuganathan and Earle, 1991; Beranyi and Greilhuber, 1995). A comprehensive study on angiosperm genome size variation was conducted by Soltis et al. (2003)) and Walker et al. (2005)) on nuclear DNA content and ploidy status of Atriplex halimus. Genome size in angiosperms varied from small values (0.05 pg) in Cardamine amara (Bennett and Smith, 1991) to huge (127.4 pg) in Fritillaria assyriaca (Bennett and Smith, 1976). Despite this huge range in DNA contents, the basic complement of genes required for normal growth and development appears to be essentially the same. Meanwhile, chromosomal aberrations could create structural and sequence composition differences between genomes of closely related species. Such changes often affect chromosome behaviors at meiosis resulting into divergence and evolution of species (Rees et al., 1982).
Pearl millet (Pennisetum glaucum) and Napier grass (P. purpureum) are the most economically important members of the genus Pennisetum, which consists of heterogeneous assemblage of species with varying reproductive behaviors. The basic chromosome numbers are x = 5, 7, 8, and 9 while ploidy levels ranges from diploid to octaploid (Martel et al., 1997; Techio et al., 2006). The primary pool consists of cultivated and wild diploids, the secondary pool has solely Napier grass and the tertiary pool includes many distantly related species of various ploidy levels which do not naturally interbreed with the primary pool.
Considering differences in chromosome number and ploidy status of pearl millet and napier grass, the higher ploidy level of napier grass over pearl millet has raised the question ¿which of the species has higher genome content? ¿Could higher ploidy status be linked to smaller chromosome and less genome size? To this end, using conventional cytogenetics and flow cytometry techniques we assessed the ploidy status and genomic content of the species. The aim was to determine the intra- and inter-specific variations in chromosome number and genome size of pearl millet and Napier grass accessions with the view of increasing the understanding of the relationship between chromosome number, ploidy status and nuclear DNA content as it affects species complexity and evolution.
Material and methods Plant material
Twenty-four genotypes of pearl millet collected from farmer’s field and research institutes in India and Nigeria were used for the study. Also, clones of five Napier grass genotypes; two wild and three cultivated were collected from different sources as shown in Table 1.
Table 1 Name and source of genotypes of pearl millet and Napier grass used for genomic size and ploidy analysis.
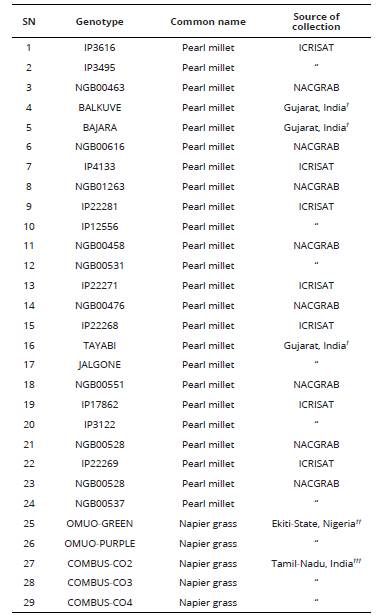
NACGRAB: National Centre for Genetic Resources and Biotechnology, Ibadan, Nigeria. ICRISAT: International Crops Research Institute for the Semi-Arid Tropics, Patanchera, Andhra Pradesh, India. † Collected from farmers’ field, in Surat District of Gujarat State, India. ††Omuo-Ekiti, Ekiti State, Nigeria. †††Department of Forage Crop, Tamil Nadu University of Agriculture, Coimbatore, India.
Plant germination and growth
The genotypes were grown in the green house at C. G Bhakta Institute of Biotechnology, Uka Tarsadia University, Bardoli, India. Two millet seedlings were raised in a planting bag while stem cutting of about two nodes were used to propagate the Napier grass genotypes. For each of the genotypes, five replicates were provided. Observations were made on reproductive behaviors and life cycle.
Chromosome number and ploidy determination
Seeds of the pearl millet genotypes were harvested from the grown plants. The seeds were germinated on moist filter paper in placed in petri dish in the Biotechnology Laboratory at CGBIBT. Root tips (0.3 - 0.5 cm) were harvested and pretreated with 8-hydroxyquinoline (2 mM) for 4 hours at 4 oC. The pretreated roots were fixed in ethanol acetic acid (3:1). Feulgen staining and squashing methods were carried out as described by Khalfallah et al. (1993)). Chromosome number and ploidy level were determined by viewing the slides under light microscope (Labophot, Nikon Corp., Japan) mounted with digital camera (Nikon DXM1200, Nikon Corp, Japan).
Flow Cytometry Analysis
Flow Cytometry determination of the nuclear DNA and genomic ploidy status was carried out using fresh young leaf tissues. About 20 mg of the leaf tissues were taken from expanding leaves for each of the genotypes of pearl millet and Napier grass. Pisum sativum (2C DNA = 8.76 pg and DNA diploid) grown under greenhouse conditions was used as an internal standard (Dolezel et al., 1998; Godelle et al., 1993). We used intercalating Propidium iodide (PI) as fluorescence stain for the genomic size determination and base specific mithramycin (MI) dye for GC % determination (Dolezel et al., 1992; Crissman and Steinkamp, 1993). The leaf tissues of each genotype were chopped and finely diced with a razor blade in a petri dish containing 0.5 ml of nuclei extraction buffer. The extraction process followed the method described by Rounsaville and Ranney (2010)). The resultant mixture was filtered into a small test tube. A solution consisting 2 ml staining buffer, 6 μl RNase A and 12 μl PI (Cystain PI Absolute P Nuclei Extraction Buffer; Partec GMBH, Munster, Germany) was added to the filtrate to give the final PI and RNase concentrations of 50 μg μl-1 and 17.5 μgl-1, respectively. The sample solutions were stored at 4 oC for 1 hour.
FACS Calibur (Becton and Dicknson, San Jose USA) Flow Cytometer was used for the analysis. A minimum of 2000 counts was carried out for each sample to estimate the nuclei DNA by internal standard method (Dolezel, 1997). Analysis were conducted in five replicates. The DNA was estimated by using the ratio of the pearl millet/Pisum sativum and Napier grass/ Pisum G1 positions, respectively, and the coefficients of variation observed for the peak within each histogram were recorded. Haploid, 2C genome size was calculated as: 2C = genome size of standard multiply by mean fluorescence value of sample in relation to mean fluorescence of the standard. The mean DNA content per chromosome was estimated as: DNA content in 2C nucleus (pg)/total chromosome number to compare the genome size at chromosome level for the genotypes. The two complementary estimates of base-composition of each of the genotype relative to a standard of known genome size and base composition (Pisum sativum; 40.5 GC %) was determined by the method of Godelle et al. (1993)). The nuclear DNA was analyzed using Cell Quest Pro Software and the result compared and validated by FloJo, a third party software.
Results
Cytological results showed that pearl millet genotypes were diploid with 14 chromosomes (2n = 2x =14). All the Napier grass genotypes were tetraploid with 28 chromosomes (2n = 28) except COM-CO-3 (2n = 21) (Figure 1). Observations on the reproductive behavior of the genotypes showed that all pearl millet were sexual, flower and fruit bearing. Napier grass genotypes Omuo green, COM-CO-3 and COM-CO-4 flowered while Omuo purple, and COM-CO-3 Napier grass did not flower during the period of the experiment. All pearl millet genotypes were annual with a life cycle of 2-3 months. The Napier grass on the other hand were perennial (Table 2).
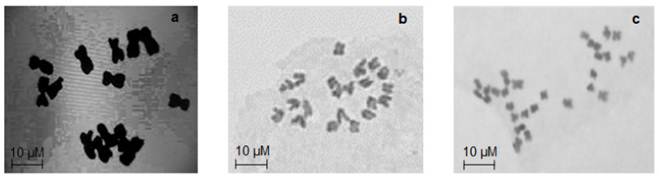
Figure 1 (a): 2n = 14 chromosome numbers of all genotypes of pearl millet; (b): 2n = 21 chromosome numbers of Napier grass COM-CO3 genotype; (c): 2n = 28 chromosome numbers of Napier grass genotypes Omuo-green, Omuo-purple, COM-CO-2 and COM-CO-4.
Table 2 DNA amount and base composition of pearl millet and Napier grass genotypes determined by flow cytometry.
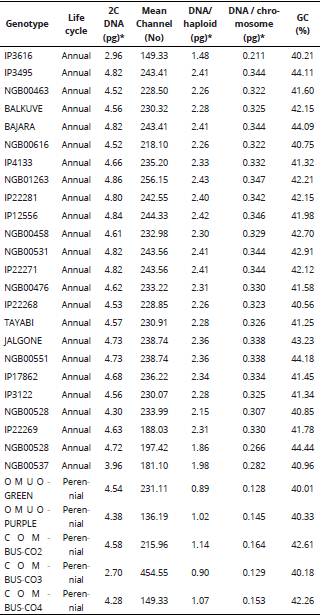
2n = number of chromosomes, 2x = diploid, 3x = triploid, 4x = tetraploid, 2C is the total genome in the accessions and %GC stands for percentage richness of Guanine and Cytosine base in the genome. *1 pg = 987 Mbp.
The DNA content per nucleus ranged from 2.96-4.86 pg and 2.70-4.58 pg for pearl millet and Napier grass genotypes, respectively. The highest amount of DNA (4.86 pg) was found in pearl millet NGB01263 genotype and the least value (2.96 pg) was recorded in IP3616 which is an ICRISAT genotype (Table 2). Among the Napier grass, the highest DNA content was found in COM-CO-2, while COM-CO-3 contained the least amount of DNA (2.70 pg) per nucleus. The mean DNA content per haploid set of genome varied, genotypes with higher ploidy levels (triploid and tetraploid) had less amount of DNA per haploid set. Similarly, the higher the ploidy status, the less the amount of DNA per chromosome.
The base composition (%GC) for the genotypes ranged from 40.01 to 44.44% (Table 2), the genomic size was directly proportional to the %GC composition. With respect to DNA ploidy level, all the studied genotypes had mean channel values less than 300 in comparison with the internal standard (Pisum sativum) which had >400 as against 156.99 - 248.66 among the pearl millet and Napier grass genotypes. Histograms of fluorescence intensities and peaks associated with nuclei isolated from leaves of pearl millet and Napier grass genotypes by flow cytometry analysis is shown in Figure 2. The peaks of the histograms marked the properties and sorting of events associated with nuclei counts of the cells from leaves of pearl millet and Napier grass in the determination of the genomic ploidy analysis by the flow cytometry procedure.
Discussion
In the present study, we observed a consistent chromosome number of 2n = 14 among the pearl millet genotypes irrespective of their sources while Napier grass had 28 chromosomes with the exception of COM-CO-3 (2n = 21). However, despite variations in the chromosome number, basic chromosome number of x =7 was maintained for the species. Earlier reports have indicated that basic chromosome number in Pennisetum varied from x = 5, x = 7, x = 8 and x = 9 (Harlan and de Wet, 1971, Morakinyo and Adebola, 1991; Techio et al., 2006). Pearl millet and Napier grass belong to the group of x=7, thus our findings agreed with the earlier reports. Cytological observation showed fewer chromosome number in pearl millet, but the chromosome sizes are larger than those of Napier grass. The fairly large chromosomes of the studied genotypes is in tandem with Jauhar (1981)), who noted that generally the x =7 members of the genus Pennisetum have larger chromosome sizes compared with other groups.
Ployploidization is a significant phenomenon that can play a role in genome rearrangement, adaptation, reproductive isolation and ultimately speciation. It is important in plant breeding because it can influence hybridization, fertility and gene expression (Adam and Wendel, 2005). The triploid Napier grass genotype is most likely a product of hybridization between pearl millet and Napier grass as elucidated by Dowling et al. (2014)). According to Martel et al. (1997)), although strong reproductive barriers exist between pearl millet and Napier grass, triploid hybrids that are partially sterile do occur in nature. Due to their allogamy nature, hybridization between the species produced AA’B genome during meiosis in such hybrids, i.e. the A’ and A genomes of Napier grass and pearl millet associate (Jauhar and Hanna, 1998; Dowling et al., 2014). Partial sterility of the resultant hybrid could therefore be attributed to reduction in paring of smaller A’ chromosomes of Napier grass with the larger A chromosomes of pearl millet. Since members of A and B genomes do not pair with one another, the B genome members remained univalent lagging behind the other chromosomes during anaphase I and II and may not be included in the developing nuclei, thus resulting in sterility (Jauhar, 1968).
Although the genomic size varied among the genotypes, the DNA contents of the pearl millet genotypes were close. Martel et al. (1997)) reported DNA content of 4.71 and 4.59 pg for pearl millet and Napier grass, respectively, which is congruent to our present findings. Whereas the previous report was exclusively based on representative genotypes of the genus, a larger number of genotypes of pearl millet and Napier grass were sampled in our study. Our observations lend credence to hypotheses of genome constitution of Napier grass propounded by Jauhar (1981)) and reinforced the allotetraploid origin of Napier grass by amphiploid mechanism through unreduced gamete and consequently decrease in chromosome size. Such decreases in chromosome size in our view, undoubtedly led to reduced genome per chromosome. Averagely, the amount of DNA in the Napier grass is similar, despite the difference in chromosome number and ploidy levels. Apparently, the genotypes of pearl millet which are cultivated species must have evolved from a common ancestor but ecologically isolated by domestication and human agricultural activities which invariably accounts for different genomic contents observed.
Alternatively, in the opinion of Martel et al. (1997)), Napier grass likely evolved from the fragmentation of large chromosomes of a diploid hypothetic ancestor (2n = 14). The variation in nuclear DNA among the polyploid Napier grass may reflect their independent evolution from diploid species of primary gene pool via auto or allopolyploidization as opined by Soltis and Soltis (2000)). Different B>’ chromosome number may contribute to variations in DNA content and compromise fertility of interspecific hybrids due to chromosomal sterility. Earlier, difference in nuclear DNA content was suggested to confer adaptation to different environmental conditions (Walbot and Cullies, 1985). In contrast, Walker et al. (2005)), noted no significant differences in nuclear DNA content of Atriplex halimus grown under different temperatures and humidity. According to Price et al. (2000)), the presence of secondary compounds in the plant tissues can affect nuclear DNA content directly or interfere with the staining and/or the fluorescence during analysis procedure.
The amount of DNA per chromosome was reduced among the Napier grass genotypes. This is likely due to decrease in chromosome size although chromosome number and ploidy level increase, without corresponding increase in the genome content. Thus, reduction in chromosome size could lead to reduced DNA content per haploid set of genome, this statement may be relative and true for the genotypes evaluated.
Sometimes, genomic size may vary independent of chromosome number. For instance, Walker et al. (2005)) observed that some tetraploid populations of Atripex halimus have more than double the diploid amount of DNA which may be due to amplification of retrotransposons, accumulation of retro-elements (Bennetzen and Kellogg, 1997) or gain of restriction fragments (Song et al., 1995). The existence of different ploidy levels in Napier grass could confer a greater genetic heterozygosity at both individual and population levels and this may enhance selective advantage in unsuitable environments. As demonstrated by Walker et al. (2005), remarkable environmental tolerance in polyploid members of the Artiplex halimus was due to their greater heterozygosity.
Among flowering plants, chromosome number and genome size varies tremendously and there is a considerable speculation regarding both the original genome size and base chromosome number of the angiosperms (Arumuganathan and Earle, 1991). It was proposed that the original base chromosome number and genome size for monocot was small (Soltis et al., 2003). From the histograms of C-values for about 2802 species constructed by Leitch et al. (1998)), species with C-values of 1.4 pg and 3.5 pg were designated as very small and small genomes, respectively. Consequently, since the pearl millet genotypes in the present study had less than 3.5 pg C-values and therefore belong to the category of small genome plants while the Napier grass (C-value < 1.4 pg) has very small genome per haploid chromosome. The small C-values we obtained for Napier grass genotypes suggest an evolutionary lineage that have experienced decrease in genome size over the years.
Genome size and base composition are directly proportional and genotypes with higher genome size are also richer in the %GC. The %GC obtained here was relatively similar and concurred with the report of Martel et al. (1997)). Meanwhile, the base composition between 40.21 43.23% we reported for pearl millet genotypes is less than the values documented for its wild relatives P. mollissium (46.5%) and P. viola (47.2%) by Martel et al. (1997). Variation in base composition have been noted in some plant species (Cerbah et al., 1995; Godelle et al., 1993). It is important to mention that variation in base composition did not provide concise information of genome sequence that could have changed in the evolutionary process. In addition, the relationship between base composition richness and genomic size may not always be linear with respect to other species. There is a correlation between mean channel number of the nuclei analysis and DNA ploidy status. The channel number obtained in this study for all the assessed genotypes was less than 300, thus indicated that all the genotypes are nuclear haploid because, channel number approximately 400 and above signifies diploid species as obtained in the internal standard Pisum satium.
Conclusion
The understanding of nuclear genome and its components could provide clues to the mechanisms that could be responsible for genome increase or decrease in the evolutionary pathway. In the present study, the difference in C-values of genotypes of pearl millet was not remarkable, but genome size of pearl millet was slightly larger than Napier grass despite the higher chromosome number and ploidy levels of the latter. The bigger chromosome size in pearl millet possibly accounts for its higher DNA content per haploid set even though the chromosomes are fewer than in Napier grass. It was possible that in the course of polyploidy evolution of the Napier grass from its hypothetical ancestor, genomic materials are lost and the evolved smaller chromosome size had reduced genome content. The information provided by the genomic and ploidy analysis of these two most economically important members of Pennisetum will enrich the existing knowledge of their genetics and enhance development of improved cultivars.















