Introduction
Colorectal cancer (CRC) is one of the leading causes of morbimortality worldwide.1 According to demographic and time projections, its global incidence is expected to increase by 60% by 2030, resulting in more than 2.2 million new cases and 1.1 million deaths per year.2 The pathogenesis of this type of cancer is complex and is not yet fully understood. However, genetic factors reportedly play a critical role in tumorigenesis.3 In this regard, Kolligs 4 reported that up to one third of the risk of developing CRC can be attributed to hereditary factors. Likewise, people with a family history of CRC are at a higher risk of developing it. Overall, genetic mutations are critical in the development of CRC and several genes and signaling pathways have been associated with its occurrence, including KRAS, BRAF, PIK3CA, RAS-RAF-MAPKand PI3K-PTEN-AKT.5-7
The main therapeutic approach in CRC patients includes surgery and subsequent chemotherapy, and it has been described that over 75% of cases that receive this treatment can be effectively cured.8 However, it has also been reported than more than 30% of these patients could develop new neoplastic polyps,8 suggesting that this treatment is not completely effective.9 Thus, new therapies have been developed, such as anti-epidermal growth factor receptor (anti-EGFR) monoclonal antibodies, considering that EGFR is the major therapeutic target in CRC.10
However, it has been described that the efficacy of anti-EGFR therapy is affected when there are mutations in KRAS, BRAF and PIK3CA genes.11,12 Therefore, these genes constitute important biomarkers in CRC, both in terms of diagnosis and prognosis. In addition, it has been reported that mutations in AXIN2 gene associated with tooth agenesis could act as a diagnostic biomarker for CRC, so the presence of variants in this gene and of this congenital developmental anomaly of the oral cavity have been proposed as predictive factors of this type of cancer.13
Non-syndromic tooth agenesis, the most common human malformation,14 is the congenital absence of one or more permanent teeth due to alterations occurring during early stages of dental development.15 In addition, an association between mutations in AXIN2 gene and teeth development anomalies has been reported in mice, suggesting their possible participation in human dental development.16 In this sense, Lammi et al.16 found that a nonsense mutation (p.Arg656Stop) in this gene was associated with the occurrence of tooth agenesis (oligodontia) and predisposition to CRC. Likewise, Rosales-Reynoso et al.17 reported that patients with the homozygous T/T genotype of the single nucleotide polymorphism rs2240308 in the AXIN2 gene have a higher risk of CRC.
Taking this information into account, adequate knowledge about the prevalence of these genetic mutations and their possible association with clinical biomarkers such as tooth agenesis by health professionals is of great importance to improve the average time to diagnosis of CRC and, therefore, the prognosis of these patients, particularly dentists, who have an important role in detecting tooth agenesis. Thus, the aim of this study was to evaluate the available scientific evidence on the prevalence of mutations in genes KRAS, PIK3CA, BRAF, AXIN2 mutations and their possible association with dental agenesis in people with CRC.
Materials and methods
We conducted a systematic review following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.18
Search strategy
A structured and systematic search using MeSH and DeCS terms was performed in Medline (Via PubMed), EMBASE (Via Ovid) and Cochrane Library databases using the following search strategy: type of studies: observational studies reporting the prevalence of KRAS, PIK3CA, BRAF and AXIN2 mutations in people diagnosed with CRC and their possible association with dental agenesis; publication period: January 2010-September 2020; publication languages: English and Spanish; search terms: "Genes", "RAS", "Kras", "PIK3CA", "BRAF", "AXIN2", "Prevalence", "Mutation", "Polymorphism", "Colorectal Neoplasms", "Colorectal Cancer" and "dental agenesis", used in different combinations ("AND" and "OR"). The search equation used in each database is shown in Appendix 1.
Screening and selection process
The titles and abstracts of the records retrieved in the searches were managed using the reference manager software EndNote® (Version X8, Thomson Reuters). After removing duplicates, two reviewers (FS and FG) independently screened all titles/abstracts to exclude those studies that were not relevant for the objective of this systematic review. Then, the full texts of the screened articles were read by the two reviewers to confirm if they addressed the topics of interest for this review and make a decision on their final inclusion for full analysis taking into account the following inclusion criterion: being case-control, cohort or cross-sectional studies addressing the prevalence of KRAS, PIK3CA, BRAF and AXIN2 mutations/polymorphisms in people with primary (adenocarcinoma) or metastatic CRC and their possible association with tooth agenesis. In addition, studies conducted in animals, those published before 2010, and those addressing other types of genetic alterations and reporting other associations with CRC or in concomitance with other cancers were excluded.
Disagreements were resolved by consensus, and when necessary, a third reviewer (MM) participated in the discussion until an agreement was reached.
Methodological quality assessment
The methodological quality of the selected studies was evaluated independently by two appraisers following criteria previously reported.19 The following criteria were evaluated: a) research question/aim of the research (1 item); b) participants (5 items); c) comparability between groups studied (4 items); d) definition and measurement of the main variables (4 items); e) statistical analysis and confounders (4 items), global assessment of internal validity; f) results (4 items); g) conclusions, external validity and applicability of results (4 items), and h) conflicts of interest (1 item). Each item was assessed using the following options: "very good", "good", "regular", "bad", "not reported" and "does not apply". Regarding the global assessment of the quality of the studies, "high", "medium" or "low" categories were used.
Two authors (FS and FG) established a grading system by assigning "5", "4", "3", "2", "1" and "0" points to the "very good", "good", "regular", "bad", "not reported" and "does not apply" assessment criteria, with 135 and 27 being the maximum and minimum scores, respectively. Studies with a score between 81 and 107 and with regular internal validity were classified as having "medium" methodological quality, while those with a score >108, as having a "high" methodological quality.
Finally, it should be noted that in those studies in which the assessment of the "comparability between groups studied" criteria was not possible due to the study design, the maximum and minimum scores were 115and 23, respectively. In these cases, studies with regular internal validity and with a score between 69 and 91 and those with a score >92 were considered to have "medium" and "high" methodological quality, respectively.
Data extraction and analysis
The following information was extracted for each study: first author, publication year, geographic region in which the study was conducted, sample size, general prevalence of the mutation, mutation prevalence by sex (male and female), mutation (changes in amino acids) and sequencing techniques. The information was included in tables and organized by genes.
Results
Selection and characterization of studies
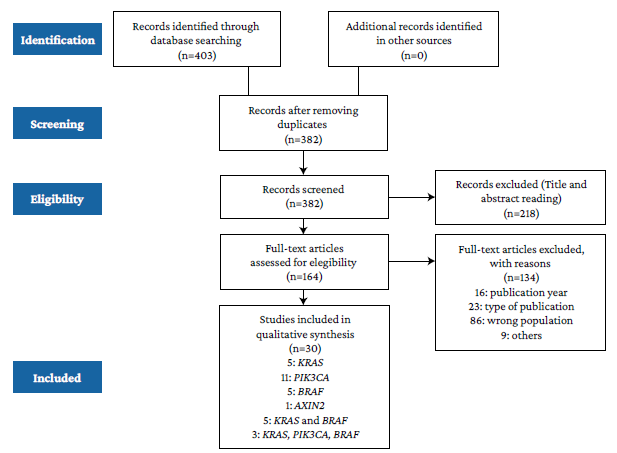
The study selection process is presented in Figure 1. In total, 30 articles were included for full analysis.
Regarding their geographical distribution, 10 studies were conducted in Asia; 8, in America; 5, in the Middle East; 3, in Europe; 2, in Oceania, and 1, in Africa. Regarding sample size, studies addressing KRAS, PIK3CA and BRAF mutations were conducted in samples ranging from 49 to 5 732, 61 to 2 299, and 17 to 1 110 individuals, respectively. Besides, all studies were cross-sectional (Table 1).
Table 1 General characteristics of the studies included for full analysis.
| Authors (year) | Country | Study design | Sample | Genes assessed | MQA |
|---|---|---|---|---|---|
| Gavin et al. 20 (2012) | United States | CSS | 2 299 | PIK3CA | High |
| Palomba et al. 21 (2012) | Italy | CSS | 384 | PIK3CA | High |
| Mao et al. 22 (2012) | China | CSS | 61 | PIK3CA | High |
| Liao et al. 23 (2012) | United States | CSS | 1 170 | PIK3CA | High |
| Watanabe et al. 24 (2013) | Japan | CSS | 5 732 | KRAS | High |
| Shen et al. 25 (2013) | China | CSS | 674 | KRAS, BRAF | High |
| Patil et al. 26 (2013) | India | CSS | 1 323 | KRAS | High |
| Chang et al. 21 (2013) | Taiwan | CSS | 165 | KRAS, BRAF | Medium |
| Rosty et al. 28 (2013) | Australia | CSS | 757 | PIK3CA | High |
| Kang et al. 29 (2013) | United States | CSS | 150 | PIK3CA | Medium |
| Marchoudi et al. 30 (2013) | Morocco | CSS | 92 | BRAF | High |
| Samadder et al. 31 (2013) | United States | CSS | 563 | BRAF | Medium |
| Baskin et al. 32 (2014) | Turkey | CSS | 49 | KRAS | Medium |
| Imamura et al. 33 (2014) | United States | CSS | 1 267 | KRAS | High |
| Bader & Ismail34 (2014) | Saudi Arabia | CSS | 83 | KRAS | High |
| Chen et al. 35 (2014) | China | CSS | 214 | KRAS, PIK3CA, BRAF | High |
| Bisht et al. 36 (2014) | India | CSS | 204 | PIK3CA | High |
| Russo et al. 37 (2014) | United States | CSS | 222 | PIK3CA | High |
| Siraj et al. 38 (2014) | Saudi Arabia | CSS | 757 | BRAF | High |
| Ye et al. 39 (2015) | China | CSS | 535 | KRAS, BRAF | Medium |
| Zhang et al. 7 (2015) | China | CSS | 1 110 | KRAS, PIK3CA, BRAF | High |
| Phipps et al. 40 (2015) | United States, Canada, Australia | CSS | 377 | PIK3CA | High |
| Foltran et al.41 (2015) | Italy | CSS | 194 | PIK3CA | High |
| Allard et al.42 (2015) | France | CSS | 1 428 | BRAF | High |
| Vatandoust et al. 43 (2016) | Australia | CSS | 3 318 | KRAS, BRAF | Medium |
| Watson et al. 44 (2016) | United States | CSS | 447 | KRAS, BRAF | High |
| Al-Shamsi et al. 45 (2016) | Arab countries | CSS | 99 | KRAS, PIK3CA, BRAF | High |
| Molaei et al. 46 (2016) | Iran | CSS | 85 | BRAF | Medium |
| Jauhri et al.47 (2017) | India | CSS | 112 | PIK3CA | High |
| Chang et al.48 (2020) | Taiwan | CSS | 161 | AXIN2 | High |
MQA: Methodological quality assessment; CSS: Cross-sectional study.
Source: Own elaboration.
Methodological quality assessment
Based on the scores obtained using the critical appraisal of epidemiological cross-sectional studies instrument, it was determined that 23 studies had "high" methodological quality, while the remaining 7 studies had "medium" quality (Table 1). The domains with the highest mean scores were "participants" and "results", whereas the lowest mean scores were observed in the "statistical analysis and confounders" domain.
KRAS
The highest overall prevalence of mutations in this gene was 54% in a sample of 447 individuals and the lowest, 20.5% in a sample of 1 323 people. The highest prevalence by sex was 65.7% and 50% in males and females, respectively. Moreover, in 46.15% and 23.08% of the studies, direct sequencing and next-generation sequencing (NGS) were carried out to identify mutations. The most frequent mutation consisted of an amino acid change from glycine into aspartic acid in codon 12 (Table 2).
PIK3CA
The highest general prevalence of mutations in PIK3CA was 20.2% in a sample of 2 299 people and the lowest, 3.5% in a sample of 1 110 individuals. In addition, the highest prevalence of mutations in men was 65.4%, and in women, 50%. Mutations were identified by means of direct sequencing and NGS in 57.14% and 21.43% of the studies, respectively. The most frequent mutation was the substitution of glutamic acid by lysine in codon 545 (p.Glu545Lys) (Table 2).
Table 2. Prevalence of mutations by gene and sequencing technique reported by the studies included in the review.
| Gene | Author (year) | Prevalence of mutations (%) | Most frequent mutation (%) | Sequencing technique | ||
|---|---|---|---|---|---|---|
| General (%) | Sex (%) | |||||
| Male | Female | |||||
| KRAS | Watanabe et al. 24 (2013) | 37.6 | 21.4 | 16.1 | NR | DS, LA |
| Shen et al. 25 (2013) | 35.9 | 19.0 | 16.5 | G12D (13.6) | DS | |
| Patil et al. 26 (2013) | 20.5 | 13.8 | 6.6 | G12A (36.5) | DS | |
| Chang et al. 27 (2013) | 35.7 | NR | NR | G12D (35.5) | PEA | |
| Baskin et al. 32 (2014) | 32.1 | 20.2 | 14.2 | G12D (12.2) | ARMS | |
| Imamura et al. 33 (2014) | 39.8 | 19.8 | 20.0 | G12D (12) | PS | |
| Bader & Ismail34 (2014) | 42.2 | 27 | 14 | G12D (45.7) | AA | |
| Chen et al. 35 (2014) | 44.9 | 25.4 | 19.8 | G12D (35.4) | DS | |
| Ye et al. 39 (2015) | 37.9 | 20.6 | 17.3 | G12D (18.4) | DS, ARMS | |
| Zhang et al. 7 (2015) | 45.4 | 25.7 | 19.6 | G12D (40.7) | DS, ARMS, NGS | |
| Vatandoust et al. 43 (2016) | 38.9 | NR | NR | NR | NR | |
| Watson et al. 44 (2016) | 54 | NR | NR | NR | PS, NGS, SNuPE | |
| Al-Shamsi et al. 45 (2016) | 44.4 | 25.2 | 19.1 | NR | NGS | |
| PIK3CA | Gavin et al. 20 (2012) | 20.1 | 10.4 | 9.7 | NR | AA |
| Palomba et al. 21 (2012) | 17.4 | 9.8 | 7.5 | E545A (14) | DS | |
| Mao et al. 22 (2012) | 8.2 | 4.9 | 3.2 | H1047L (7) | DS | |
| Liao et al. 23 (2012) | 16 | 8.2 | 7.8 | NR | PS | |
| Rosty et al. 28 (2013) | 14 | 6.8 | 7.2NR | E542K (35) | DS | |
| Kang et al. 29 (2013) | 12 | NR | NR | NR | DS | |
| Chen et al. 35 (2014) | 12.3 | 8.0 | 4.2. | H1047R (31) | DS | |
| Bisht et al. 36 (2014) | 5.9 | 2.9 | 2.9 | E545K (3.4) | DS | |
| Russo et al. 37 (2014) | 13 | NR | NR | NR | DS | |
| Zhang et al. 7 (2015) | 3.5 | 1.9 | 1.5 | H1047R (3.5) | DS, ARMS, NGS | |
| Phipps et al. 40 (2015) | 11 | 4.5 | 6.6 | E542K,E545K (64) | PS | |
| Foltran et al. 41 (2015) | 16.49 | NR | NR | E545K (56) | PS | |
| Al-Shamsi et al. 45 (2016) | 13.1 | 5.1 | 8.08 | NR | NGS | |
| Jauhri et al. 47 (2017) | 16.1 | 13.3 | 2.6 | E545A, E545K, H1047R (15.8) | NGS | |
| BRAF | Chang et al. 27 (2013) | 4.24 | NR | NR | V600E (100) | HRM |
| Shen et al. 25 (2013) | 6.96 | 4.1 | 2.8 | V600E (1.8) | DS | |
| Marchoudi et al. 30 (2013) | 5.4 | NR | NR | V600E (100) | DS | |
| Samadder et al. 31 (2013) | 27 | NR | NR | V600E (100) | DS | |
| Chen et al. 35 (2014) | 4.2 | 2.3 | 1.8 | V600E (89) | DS | |
| Siraj et al. 38 (2014) | 2.5 | 1.4 | 1.0 | V600E (89.5) | DS | |
| Allard et al. 42 (2015) | 6.4 | NR | NR | V600E (100) | HRM, DS | |
| Zhang et al. 7 (2015) | 3.1 | 1.6 | 1.4 | V600E (100) | DS, ARMS, NGS | |
| Ye et al. 39 (2015) | 4.4 | 1.5 | 2.8 | V600E (80) | DS, ARMS | |
| Vatandoust et al. 43 (2016) | 12.1 | NR | NR | NR | NR | |
| Al-Shamsi et al. 45 (2016) | 4.0 | 2.0 | 2.0 | NR | NGS | |
| Molaei et al. 46 (2016) | 0 | 0 | 0 | NR | DS | |
| Watson et al. 44 (2016) | 0 | 0 | 0 | NR | PS, NGS | |
| AXIN2 | Chang et al. 48 (2020) | 21.7 | NR | NR | A603P (11.4) | NGS |
DS: Direct sequencing; LA: Luminex assay; PEA: Primer extension assay; ARMS: Amplification refractory mutations system-PCR, PS: Pyrosequencing; AA: Array analysis; NGS: Next-generation sequencing; NR: Not reported; SNuPE: Single-nucleotide primer extension; HRM: High resolution melting.
Source: Own elaboration.
BRAF
The highest overall prevalence of mutations was 27% in a sample of 563 individuals and the lowest, 2.5% in a sample of 757 individuals. Besides, it is worth noting that BRAF mutations were not identified in two studies. The highest prevalence in men was 55.6%, and in women, 44.4%. Mutations were detected using direct sequencing and NGS in 69.23% and 23.08% of the studies, respectively. The most frequent variant was an amino-acid change of valine by glutamate in codon 600 (p.Val600EGlu) (Table 2).
Discussion
The fact that cancer cells have multiple genetic mutations suggests that the development and progression of tumors could be partially caused by mutagenesis. Additionally, these mutations can contribute to developing resistance to conventional oncological therapies, such as chemotherapy.49 Currently, scientific evidence shows that, despite the development of new drugs, therapy against cancer is limited, since new ways of resistance have emerged, such as drug inactivation, alteration of drug targets, drug efflux, and cell death inhibition.50
Taking the above into account, understanding the distribution of mutations in oncogenes in cancer patients is essential to improve both the knowledge of the genomic profile of malignant diseases and personalized medicine, since a better understanding of the cancer genome is important to choose the best treatment for each oncological patient according to their individual characteristics.51
Several studies have reported the presence of mutations in multiple genes involved in the development and progression of CRC, being KRAS, PIK3CA and BRAF the ones with the highest prevalence of mutations.6,7In addition, AXIN2 has been identified as a potentially useful gene in the early diagnosis of CRC through clinical markers such tooth agenesis.16
Regarding KRAS gene, it has been described that mutations in this gene are the mutations most frequently identified in the development of human tumors (approximately 30%).52,53KRAS encodes for a protein constituted by 188 residues of amino acids involved in molecular pathways activation that allows signal transduction from the cell surface to the nucleus.54KRAS is found in chromosome 12 and is a member of the RAS gene family; mutations in this gene comprise 86% of all RAS family mutations, being most frequently observed in codons 12 and 13 of exon 2,55 and less frequently, in codons 6156 and 146.57,58 In addition, the main mutation in KRAS consists of a G>A transition followed by a G>T transversion in exon 1.59
KRAS has been studied as a predictive molecular marker of anti-EGFR monoclonal antibody resistance in primary and metastatic CRC patients.11,60,61 In the presence of KRAS mutations, GTPase activity decreases and the KRAS mutant protein remains bound to GTP in its active conformation, transmitting signals continuously. As a result, signal transmission is not blocked by EGFR inhibitors and the effects of this therapy are scarce or cannot be observed.62-64
In the present review, the prevalence of KRAS mutations ranged between 20.5% to 54.7,24-27,32-35,39,43-45 Regarding the prevalence of these mutations in different regions of the world, the following was found:
In America, the highest prevalence was 54% in a sample of 447 individuals,44 while the lowest was 40% in a sample of 1 267 people.33 In relation to sex distribution, the highest prevalence of mutations in males was 19.8, while in females it was 20%.33
In Asia, the highest prevalence of KRAS mutations was found in China (45.4% in a total sample of 1 110 individuals),7 while the lowest was found in India (20.5% in a sample of 1 323 people.26 Regarding sex distribution, the highest prevalence of mutations in males was 25.7%,7 while in females it was 19.8%.35
In the Middle East, the highest prevalence of KRAS mutations was 44.4% in a sample of 99 individuals from several countries,45 and the lowest prevalence was 30.6% in a sample of 49 people from Turkey.32 In addition, the highest prevalence of mutations in males was 27%,34 while in females it was 19.1%.45
Only one study included in this review provided data on KRAS mutations in Oceania, reporting a prevalence of 38.9% in a sample of 3 318 Australian subjects; however, no data on prevalence by sex were reported.43
Finally, the most frequent mutation in the KRAS gene was the substitution of glycine for aspartate (p.Gly12Asp). However, the substitution of glycine by alanine (p.Gly12Ala) was the most common mutation in one study.26 These conformational biochemical changes have been associated with a poor prognosis in terms of survival and with increased tumoral aggressiveness.32,65,66
PIK3CA is a gene located in chromosome 3 that codes for PI3K protein. PI3K is part of the lipid kinase family, which is involved in cell proliferation, growth and survival.67,68PI3K protein is also involved in the PI3K/AKT pathway, which catalyzes AKT phosphorylation, activating the downstream signaling pathway.69
It has been reported that the presence of mutations in this gene stimulates said pathway and promotes cell growth in various types of cancers.70 The prevalence of mutations in the PIK3CA gene ranges from 7% to 32% in CRC patients, being the G>A transversion in exons 9 and 20 the most common mutation (80% of mutations). Furthermore, there is contradictory evidence in relation to the presence of these mutations as a predictor of response to cancer treatment. 28,71,72 On the one hand, mutations in exon 20 have been associated with a low response to treatment with cetuximab and chemotherapy.73 On the other, clinical trials such as the one conducted by Soeda et al.74 state that their presence might not contribute to the prediction of the response to monoclonal therapy with cetuximab in patients with advanced and/or metastatic CRC.
According to the data retrieved in the present systematic review, the prevalence of mutations in PIK3CA ranged between 3.5% to 20.1%.7,20-23,28,29,35,36,37,40,41,45,47 Regarding the prevalence of these mutations in different regions of the world, the following was found:
In America, the highest prevalence was 20.2%, in a sample of 2 299 individuals from the United States,20 while the lowest prevalence was 11%, in a sample of 377 people from the United States and Canada.40 In addition, the highest prevalence of mutations in males was 10.4% and in females, 9.7%.20
In Asia, the highest prevalence of PIK3CA mutations was reported in India (16.1% in a sample of 112 patients),47 while the lowest was found in China (3.5% in a sample of 1 110 people).7 In addition, the highest prevalence of mutations in males was 13.3%47, and in females, 4.2%.35
Only one study reported data for people from the Middle East, finding a prevalence of PIK3CA mutations of 13.1% (5.1. in males and 8% in females) in 99 individuals from Middle Eastern countries.45
Similarly, only one study reported data for population from Western Europe, finding a prevalence of PIK3CA mutations of 17.4% (9.8% in males and 7.5% in females) in a sample of 384 Italian individuals.21
Only one study provided data on PIK3CA mutations in Oceania, reporting a prevalence of 14% (6.8% in males and 7.2 in females) in a sample of 757 individuals from Australia.28
Finally, the most frequent variant in the PIK3CA gene was the replacement of glutamic acid by lysine (p.Glu545Lys) due to alterations in exon 9. However, a high mutation index was also reported in exon 20, resulting in the substitution of histidine by leucine (p.His1047Leu) and of histidine by arginine (p.His1047Arg).
BRAF is a gene involved in cell proliferation and differentiation, as well as in apoptosis pathways;75 besides, it has been described that the presence of mutations in in this gene might lead to phenotypic alterations in the colorectal tissue.75 In the present review, the prevalence of mutations in BRAF ranged from 2.5% to 27%.7,25,27,30,31,35,38,39,42-46Regarding the prevalence of these mutations in different regions of the world, it was found that:
In America, the highest prevalence was 27% in a sample of 563 individuals.31 In contrast, the lowest prevalence (0%) was reported by Watson et al.44 in a study conducted in 17 people; both studies were carried out in the United States. No data on the prevalence of these mutations in men and women were reported.
In Asia, the highest prevalence was 6.96% in a sample of 674 people25 and the lowest was 3.1% in a sample of 1 110 individuals,7 with both studies being conducted in China. With regard to the prevalence of BRAF mutations by sex, the highest prevalence in men was 4.1% and in women, 2.8%.25,39
In the Middle East, the highest prevalence of mutations was 4% in a sample of 99 individuals from several countries,45 while the lowest prevalence (0%) was reported by Molaei et al.46 in a study conducted in 85 people from Iran. Furthermore, the highest prevalence of mutations was 2% in both males and females.45
Only one study reported data on BRAF mutations in population from Western Europe, finding a prevalence of 6.4% in 1428 individuals from France.42 No data on the prevalence of these mutations by sex were reported.
In Africa, a study conducted in 92 people from Morocco found a prevalence of 5.4%, however no data on the prevalence of these mutations stratified by sex was reported.30
In the case of Oceania, a study conducted in 173 individuals from Australia reported a prevalence of mutations of 12.1%.43
Finally, the most frequent mutation in this gene was the substitution of valine by glutamate in codon 600 (p.Val600Glu). On the other hand, it is worth noting that Samowitz et al.76 reported that individuals with mutations in BRAF have more aggressive CRC pheno-types and show a poor response to treatment with cetuximab or panitumumab. Thus, and given that most of the studies included in this review only studied the p.Val600Glu genetic variant, further research on other possible hotspot regions in this gene associated with CRC is required.
Regarding studies conducted before 2012, the following data on the prevalence of mutations in the KRAS, PIK3CA and BRAF genes was found: Segura-Uribe et al.77 and Vaughn et al.78 reported a prevalence of KRAS mutations of 32.4% and 42.4% in 37 colorectal tumors in Mexican individuals and in 2 121 colorectal adenocarcinomas in people living in the United States, respectively. Herreros-Villanueva et al.79 and Velho et al.80 reported a prevalence of PIK3CA mutations of 8.22% and 7.1% in CRC specimens of 73 Spanish individuals and in 103 CRC carcinomas collected from Portuguese individuals, respectively. Finally, Nicolantonio et al.11, in a study conducted in 113 patients with metastatic CRC from Italy and Switzerland, reported a prevalence of BRAF mutations of 14%.
After comparing these data with those reported in the studies included in this systematic review, it is possible to say that the frequencies of mutations in KRAS, PIK3CA and BRAF genes in CRC patients have not changed much in recent years. However, systematic reviews that include a greater publication period are necessary to evaluate increasing or decreasing trends in the prevalence of mutations in these genes in CRC patients.
The differences between the general prevalence of mutations and their prevalence in men and women in the same geographical area could be attributed to both differences in the sample sizes of the studies and the sensitivity of the molecular techniques used, which have been shown to influence the frequency in which these mutations are detected.81,82Other factors that might influence said frequency include the quality and quantity of DNA obtained, tumor heterogeneity and possible environmental exposures unknown to the researchers or that they cannot control.83,84
Although in most of the studies included in our review there was not a significant association between the sex of the patient and the prevalence of mutations in KRAS, PIK3CA, BRAF and AXIN2 , said association was evaluated since it has been described that associations between genetic mutations and the sex of the individual could provide information about the pathogenesis of different diseases,85 and because of the differences, both environmental and genetic, that have been described between men and women around the world in terms of susceptibility to and incidence of cancer.86-88Therefore, it can be assumed that the differences in the prevalence of mutations in these genes between males and females might condition a higher frequency of CRC in individuals of a specific sex.
The incidence and mortality rates of CRC have increased in recent years,89 therefore, there is a higher need to identify and implement diagnostic strategies for the early detection of this cancer, including the analysis of molecular and clinical biomarkers. In this regard, it has been suggested that AXIN2 gene could be considered a molecular biomarker of CRC, since the presence of mutations in this gene and of tooth agenesis have been proposed as predictive factors of CRC.13,16
In this regard, the present systematic review aimed to report the prevalence of mutations in AXIN2 in individuals with primary and/or metastatic CRC around the world. However, only one study was retrieved. In addition, due to the knowledge of the possible associations of AXIN2 gene with both CRC phenotypes, we sought to identify if some of the most prevalent genetic mutations in CRC, such as the ones in KRAS, PIK3CA and BRAF genes, were also associated with tooth agenesis; nevertheless, no studies reporting such an association were identified.
AXIN2 is known for its tumor suppressing activity by negatively regulating the Wnt pathway trough the intracellular degradation of ß-catenin.90,91In mice, AXIN2 is expressed during odontogenesis in dental mesenchyme, enamel knot, dental papilla and mesenchymal odontoblast.16 It is reasonable to hypothesize that an impairment in this gene could affect the development of molars and incisors, leading to tooth agenesis.92 In addition, there is evidence that the expression of AXIN2 in colorectal tissue can lead to carcinomas.16 Wu et al.93 reported that mutations in AXIN2 could influence the expression of the protein it codes for, which can play a critical role in carcinogenesis, a similar claim to that of Rosales-Reynoso et al.,17 who state that these mutations act as a genetic risk factor for the development of CRC. Similarly, Marvin et al.94 reported that the presence of nonsense mutation p.Tyr663X (c.1989G>A) in AXIN2 results in protein truncation in individuals with oligodontia and gastrointestinal neoplasms. Therefore, tooth agenesis and the presence of variants in AXIN2 could be used as clinical and molecular markers for the development of CRC.
The present study highlights the importance of researching the distribution of mutations in KRAS, PIK3CA, BRAF and AXIN2 genes in people with CRC from different regions of the world to determine the impact of these variants in the early diagnosis of this type of cancer, as well as in its prognosis in terms of survival and efficacy of therapies. Studying the presence of genetic mutations in heterogeneous populations is of great importance, given the possible association between a higher probability of mortality due to CRC and the ethnic and sexual differences in the presence of certain genetic mutations. In this regard, ethnicity has been shown to be associated with a higher risk of cancer93,95-97and with a worse prognosis in the presence of KRAS,98PIK3CA29 and BRAF99 mutations.
Conducting additional studies on the association between genetic mutations, including those in KRAS, PIK3CA, BRAF and AXIN2 genes, and the development of CRC in diverse populations is recommended in order to contribute to the knowledge on the genome of this type of cancer genome. Likewise, conducting studies that analyze the association between the presence of mutations in AXIN2 and dental agenesis as a clinical marker for the early diagnosis of CRC is also recommended.
Conclusions
Our findings suggest that worldwide there is a diverse distribution of KRAS, PIK3CA and BRAF mutations in individuals with CRC, being KRAS mutations the most prevalent. Moreover, according to the evidence here retrieved there is no association between tooth agenesis and KRAS, PIK3CA and BRAF germline gene mutations in these patients. AXIN2 is the unique gene in which an association with both phenotypes (i.e., primary and metastatic) has been well established, but population studies on the prevalence of AXIN2 mutations are limited.