Introduction
Rotaviruses are the most important viral cause of diarrhea in a wide variety of domestic animals and children1,2. In cattle, it is one of the main agents involved in the development of Bovine Neonatal Diarrhea (BND), a disease that mainly affects calves in the first month of life and causes high economic losses associated primarily with costs generated by pharmacological treatments, decreased growth averages in calves, and high morbidity and mortality3 . Group A rotaviruses account for approximately 95% of rotavirus infections in cattle worldwide4.
Rotaviruses are members of the family Reoviridae. Structurally, they are icosahedral, triple-layered, and non-enveloped particles5,6. Their genome is double-stranded RNA (dsRNA) and is divided into 11 segments 1,5 that encode six structural proteins (VP1, VP2, VP3, VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6)5. Antigenic and genomic analyses of the VP6 protein have allowed the classification of rotaviruses into seven groups (A-G)7 , among which A, B, and C infect humans and animals 7, while groups D to G have been associated with illness in animals only3,7. Rotaviruses are also classified into G and P types, which are determined by the outer layer viral proteins, VP7 ( genotype G for glycoprotein) and VP4 (genotype P for protease sensitive)1,8. To date, 27 G and 35 P genotypes have been described7, of which P [1], P [3], P [5], P [7], P [11], P [14], P [17], P [21], P [29], P [33] and G1, G2, G3, G5, G6, G8, G10, G11, G15, G17, G21 and G24 [1, 3, 5, 6, 7, 8, 10, 15] are prevalent in cattle9,10. Due to the segmented nature of the rotavirus genome, it can reassort during multiple infections. As the gene segments encoding VP7 and VP4 can segregate independently during reassortment, different G and P type combinations can be found in co-circulating rotaviruses8. Among the possible combinations between genotypes G and P, only G[6], G[10] and G[8] combined with P[5], P[11] and P[1] are considered epidemiologically important in cattle11,12. Additionally, other less common combinations G21 P[29], G24 P[33] , G17P[17] have been reported with greater frequency in other species different from bovine, demonstrating the occurrence of bovine rotavirus gene reassortment9,10. On the other hand, some studies have classified the G6 genotype into five genetic lineages: type I lineage includes human rotavirus P9; type II lineage contains human and goat P14; type III lineage spans human rotavirus P9 and buffalo P3; type IV lineage includes bovine rotaviruses P1 and P5, and type V lineage contains only bovine rotavirus P1111,12.
In Colombia, the prevalence of bovine rotavirus as a causal agent of BND development13, as well as the circulation of the G6 genotype14 have been reported. The objective of this study was to characterize rotaviruses affecting calves in the Sabana region of Bogotá, Colombia, by viral genome detection and gene fragment sequencing, in order to determine homology with bovine rotaviruses isolated from Latin America and worldwide.
Materials and Methods
Fecal samples
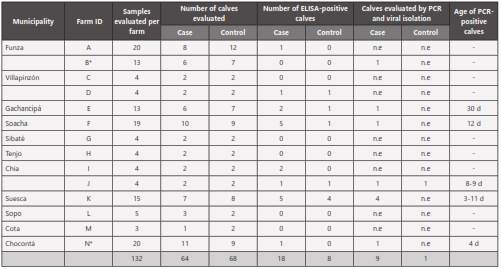
A convenience sampling was done during the first five weeks of life on 132 calves from 14 farms located in 11 municipalities of the Sabana region of Bogotá, Colombia. Of these samples, 64 were taken from calves with clinical signs of BND (cases) and 68 samples from clinically healthy calves (controls) (Table 1). The number of calves sampled per week (W) was: W1: 45; W2: 41; W3: 28; W4: 6; W5: 12.
ELISA
Fecal samples were processed using an antigen capture ELISA kit (Digestive Bio K 071®, Bio-X Diagnostics, Belgium). The sensitivity and specificity were between 77% and 97% and 90 and 100%, respectively, according to the manufacturer laboratory. Optical density (OD) reading was performed using an ELISA reader (BioStackReady® BioTek® Power Wave Reader XS) at 450 nm wavelength. The results were obtained by subtracting the absorbance value of the sample from the absorbance value of the negative control, such that samples were considered positive when the result was higher than 0.150 Elisa units (EU).
Viral isolation
A random selection was made among samples that arrived at the laboratory in less than 12 hours. A total of nine samples (eight cases and one control) were selected from five farms in five municipalities (Suesca, Chia, Gachancipá, Soacha, and Chocontá), of which two samples were taken from Chia and four from Suesca. These samples were evaluated by PCR and processed for viral isolation (Table 1). For this, the MA104 cell line was used, which was donated by Dr. Carlos Guerrero from the Virology Laboratory, Faculty of Medicine, Universidad Nacional de Colombia, Bogotá. The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% irradiated fetal bovine serum (Gibco®), 1% penicillin-streptomycin-amphotericin solution (Gibco®), and 2% glutamine (Sigma®), and maintained at 37°C with 5% CO2 . As positive control, the RF rotavirus strain, also donated by Dr. Carlos Guerrero, was used. Samples were processed from 1g of fecal matter that was subjected to homogenization in 4ml of PBS and sonication for 30 sec at 30Ω, followed by centrifugation at 4.000xg for 10min and supernatant recovery. The supernatant was incubated for 1 hour at room temperature with gentamicin at a final concentration of 0.5 mg/ml. The inoculum was activated by addition of trypsin TPCK (SigmaAldrich®) to a final concentration of 100 μg/ml, followed by incubation at 37°C for 1 hour (Elschner et al 2002**). For viral infection, a monolayer of MA104 cells in a 12-well plate was washed with PBS and pretreated with 200 μl of diethylaminoethyl (DEAE)-dextran solution (Sigma-Aldrich®) (40 mg DEAE-dextran/L DMEM) and incubated for 30 minutes at 37°C. Subsequently, the medium was removed, 100 μl of the inoculum were added, and the cells were incubated one hour at 37°C. The inocula were then replaced with DMEM containing 10 μg/ml of trypsin, 5% fetal bovine serum FBS, 1% L-glutamine, and 50 mg/ml gentamicin, and the cells were incubated at 37°C in a 5% CO2 atmosphere for 1-5 days until signs of cytopathic effect.
Detection of the viral genome
Total RNA was extracted from the cells that developed a post-infection cytopathic effect, using the Micro Prep-ZymoResearch® Quick RNA ™ Kit, following the manufacturer’s recommendations. Subsequently, cDNA synthesis was performed through RT-PCR using random primers and M-MVL reverse transcriptase (Invitrogen®). For rotavirus detection, a segment of the VP7 protein-encoding gene was amplified through a nested PCR. VP7 primers for the first PCR reaction were VP7-F 5´ GCCTTTAAAAGCGAGAATTT 3´and VP7-R 5´GGTCACATCATACAACTCTA 3´ 1 and for the second PCR reaction these were BRV-VP7Int F 5´ GTATGGTATTGAATATACCAC 3´ and BRV-VP7Int R 5´ GATCCTGTTGGCCATCC 3´ 15, which amplified regions of 1062 and 341 bp, respectively. The final reaction volume for the nested PCR was 25 μl, containing 2.5 μl 10X PCR buffer, 1.25 μl of each primer (1 μM), 0.1 μl Taq platinum-Invitrogen® (5U/ μl), 1.25 μl MgCl2 (2.5mM), 1.25 μl dNTP mix (0.5mM), and 2 μl cDNA. The PCR was done with the following conditions: pre-denaturation at 94°C for 5min, followed by 30 cycles of 94°C for 1min, 56°C for 1min and 72°C for 2min, and a final extension at 72°C for 8min. These conditions were employed for the two PCRs and were performed on a Biorad®-DNA Engine thermocycler. One microliter of the first amplification reaction was used for the nPCR. As positive control, the bovine rotavirus reference RF viral strain was used and DEPC-treated water was used as negative control. Controls were included in each PCR run, and the controls from the primary PCR was carried over to the nested step.
Sequencing
Nine PCR products for the VP7 region were Sanger sequenced by Macrogen® USA, however only 6/9 were obtained in good quality for their alignment. Dual direction sequencing was obtained from 10 μL of purified amplicon (Gel extraction purification kit, QIAGEN®). Forward and reverse sequences were assembled and edited on BioEdit 7, aligned with ClustalW (Mega 4.0.2), and used for phylogenetic analyses based on Maximum Likelihood (ML) using the Tamura 3-parameter (G+I) substitution model, selected as best-fit model according to the Bayesian Information Criterion, and 10,000 Bootstrap replicates (Mega 4.0.2).
Statistical analysis
To determine if there was a correlation between the presentation of diarrhea with age and the presence of rotavirus, a logistic model was used in which the odds of disease probability (diarrhea) were modeled according to age in weeks and the presence or absence of rotavirus. The software used was 9.2 SAS/STAT® Institute Inc., Cary, NC; and differences were considered statistically significant at p<0.05.
Ethical considerations
All procedures for this study were approved by the Ethics Committee, equivalent to the Institutional Animal Care and Use Committee (IACUC), of the Faculty of Veterinary Medicine and Animal Sciences of the Universidad Nacional Colombia.
Results
Of the 132 fecal samples, 26 (19.7%) were positive by the ELISA test, 18 corresponded to calves with diarrhea, and 8 to clinically healthy (controls) (Figure 1). The 26 rotavirus-positive calves came from 8 farms (57%) located in 7 municipalities, where a positivity of 63% was found. By ELISA, two farms (Chocontá and Soacha) were found to be positive by only one sample, while in the remaining six farms, 2 to 9 positive calves were found. The statistical analysis indicated that the model that explains the occurrence of the disease as a result of the various combinations between age in weeks and the presence or absence of rotavirus was not significant (P-value = 0.47).
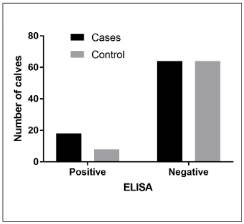
Figure 1 Distribution of ELISA-positive and negative fecal samples from calves with clinical signs of BND (cases) and clinically healthy calves (controls).
The cytopathic effect characteristic of rotavirus infection, such as cell vacuolization and monolayer detachment, was developed post-infection with all nine ELISA-positive samples. This effect was observed in different passages, 22% (2 samples) in passage 2, 33% (3 samples) in passages 3-4, and 12% (1 sample) in passage 5. The ELISA-negative calf sample did not develop a cytopathic effect.
PCR amplification of the 341bp-fragment of the VP7 gene in the nine samples that developed a cytopathic effect allowed to confirm the presence of rotaviruses (Figure 2). The ELISA-negative calf sample did not amplify by PCR.
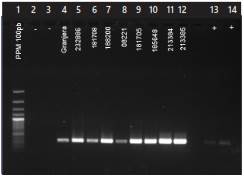
Figure 2 Nested PCR for the detection of bovine rotaviruses from cell cultures infected with calf fecal inoculum. Lanes: 1. Molecular weight marker. 2. Negative control, DEPC-treated water. 3. Negative control (ELISA-negative calf sample). 4-12 ELISA-positive samples from calves with diarrhea. 13 and 14. Positive control bovine rotavirus RF strain.
Six samples out of the nine that amplified for bovine rotavirus were sequenced. Of these, 5/6 showed homology to the G6 genotype and 1/6 to the G10 genotype. According to the classification of G6 by lineages, these viruses showed proximity to those of lineage V, corresponding exclusively to type P11 (Figure 3). No phylogenetic relationship was found with viruses of lineages III and IV, corresponding to virus types P9, P1, and P5. The sample from the calf with genotype G10 rotavirus corresponded to a clinically healthy animal (control) of two weeks of age from farm J of the municipality of Chia. In this farm, two rotavirus-positive samples were found, one from a calf with diarrhea (no genotype classified) and another from a clinically healthy calf, corresponding to genotype 10.
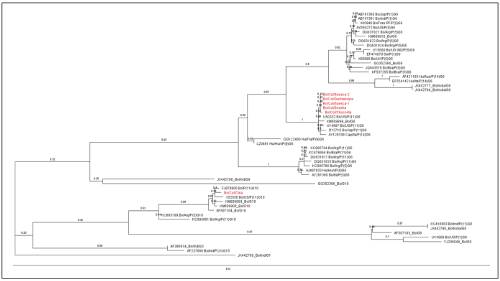
Figure 3 Phylogenetic analysis of Bovine rotavirus VP7 sequences. A Maximum Likelihood tree was constructed from aligned nucleic acid sequences of six Colombian isolates (shown in red) and 49 sequences found in GenBank (shown in black, with the GenBank accession number). Tree branch support values were calculated based on 10,000 Bootstrap replicates.
Discussion
The present study confirmed the presence of type A bovine rotaviruses in dairy herds of the Sabana region of Bogotá, Colombia. These viruses have been reported to cause 90% of infectious bovine diarrheas4 and, together with Cryptosporidium parvum, are the main infectious agents associated with BND presentation16-19.
The presence of rotavirus was found in 19.7% of the analyzed samples (n = 132) and in 28.1% of the calves that presented neonatal diarrhea (n = 64). These results are similar to other studies reporting a prevalence ranging from 17.7% to 20% of the total evaluated samples16,20. It is important to note that in the present study, we found a viral presence of 11% by ELISA in calves with no clinical signs (diarrhea), indicating that these could be virus disseminators (healthy carriers). Likewise, the presence of the virus was found in 63% of the municipalities evaluated, indicating a broad distribution of the pathogen in the Sabana region of Bogotá. It should be taken into account that the prevalence for these viruses may vary according to factors such as the time of the year and the type of population studied. Thus, Garaicoechea et al 2006 reported a prevalence between 29 and 100% for bovine rotavirus over a 10- year period, and found a higher prevalence in beef cattle (71. 3%) compared to dairy cattle (58.5%). In Colombia, previous studies report a 7% prevalence14 in the same region where the present study was done.
The nested PCR technique confirmed the presence of rotaviruses in fecal samples that were previously diagnosed by ELISA. No correlation was found between the occurrence of diarrhea with age and the presence of rotavirus. It is suggested that, since a greater number of diseased animals were found in the second week of age, there could be some correlation with age, although the amount of data gathered was not enough to prove this affirmation. Detection of the virus in the first weeks of calf life is similar to that reported by Foster and Smith 2009 21, where they determined that these viruses preferentially affect calves under three weeks of age, reaching a peak incidence at six days old.
As previously reported, genotypes G1, G2, G3, G6, G8, G10, and G11 are the most frequent in calves, among which the G6 genotype is predominant in BND around the world 10,22. In this study, six samples were sequenced, which aligned with bovine rotavirus strains of different G genotypes reported in Genbank. The sequence alignments showed five samples belonging to the G6 genotype, all corresponding to calves with diarrhea (cases), while a sample from a healthy calf grouped with the G10 genotype. Different studies around the world report the G6 genotype as the most prevalent in cattle, ranging from 56.7 to 74%, while G10 is less prevalent (16%), and G8 is found between 0.6 to 3.5%3,10,22,23. Gutiérrez and Baoming 200714 reported the presence of the G6 genotype for the same region of the country; however, this study is the first to report the presence of the G10 genotype in Colombia. On the other hand, studies by Garaicoechea et al. 2006 and Martella et al.200311,12 classified the G6 genotype into five genetic lineages: lineage I includes human rotavirus P9; lineage II contains human and goat P14; lineage III corresponds to human rotavirus P9 and buffalo P3; lineage IV includes bovine rotavirus P1 and P5; and lineage V contains only bovine rotavirus P11. By comparing the sequences used in the above mentioned publications with those obtained in the present study, we determined that the isolates belong to the G6 genotype and correspond to lineage V.
In our study, we found that all rotaviruses belonging to the G6 genotype were associated with the presence of diarrhea, while the only case of G10 genotype came from a clinically healthy animal. Based on these findings, it could be proposed that genotype is associated with the presentation of clinical signs. To prove this, it would be necessary to expand the molecular characterization of clinically healthy animals that were positive by ELISA.
The Sabana region of Bogotá comprises a group of 23 municipalities. In the present study, five samples showed 99% sequence identity. Of these, four came from three municipalities (Chocontá, Gachancipa, and Suesca) that are separated by distances between 14 and 26 km. This may demonstrate that there is not much variability among bovine rotavirus strains affecting nearby municipalities. Of the 14 farms evaluated, two use a multivalent vaccine against BND that is applied to cows at 16 weeks before delivery. The bovine rotavirus antigen present in the vaccine is inactivated and belongs to the G6 genotype. In one of these two farms (farm N), the virus was detected and classified as G6. This finding may be attributed to an immunogen management problem or that the positive calf did not receive good passive immunity. Finally, due to the nature of these viruses, including their ability to produce re-arrangements and their inter-species transmission, it is recommended that further studies be conducted to determine the circulating genotypes and whether they might have the capacity to infect the human population.