Introduction
The development of resistance to chemotherapy remains one of the main drawbacks in the treatment of different types of cancer. The results of different studies suggest that reactive oxygen species (ROS) may play an important role in the regulation of drug resistance. 1-3 In addition, some authors have proposed that ROS may also have some effect on the regulation of hypoxia-inducible factor (HIF-1α) activity, which is one of the main transcription factors involved in cell signaling under hypoxic conditions. 4-6 Likewise, high levels of ROS under normoxic condition may increase the activity of HIF-1α; however its effect on hypoxia is still controversial, as some authors suggest that it would favor the degradation of HIF-1α and others point an increase in the expression of the transcription factor induced by the presence of ROS. 7,8
Some of the mechanisms that explain the role of hypoxia in chemotherapy resistance in cancer include the dependence of many chemotherapeutic agents on the formation of free radicals derived from oxygen and the induction of HIF-1α, as these factors regulate the expression of various genes involved in tumorigenesis. 9 Hypoxia in colon cancer cells shows a decrease in the expression of pro-apoptotic proteins Bid and Bad, and HIF-1α is necessary to regulate Bid. A decrease in cellular apoptosis by chemotherapy has also been reported, suggesting that HIF-1α could be one of the factors involved in the development of hypoxia-induced chemoresistance in cancer cells. 10
Doxorubicin is an anthracycline used regularly to treat different types of cancer and its mechanism of action involves the induction of ROS. 11 Breast, lung, gastric and ovarian cancer, among others, have shown resistance; however, its use in association with hypoxic effect has not yet been evaluated in colon cancer cells. Said resistance could be explained by a regulatory mechanism of apoptotic effectors. The PUMA pro-apoptotic protein (p53 upregulated modulator of apoptosis) belongs to the pro-apoptotic BH3 protein family. This group includes PUMA, Bid, Bad, Bim and Noxa proteins, which stimulate apoptotic pathways, as do other members of the Bcl-2 family such as Bax and Bak. 12 The expression of PUMA is regulated by the p53 tumor suppressor protein 13, although PUMA is thought to be involved in the p53-dependent/-independent pathways of apoptotic cell death through different signaling pathways and is regulated by a wide arrange of transcription factors. After its activation, the PUMA protein interacts with proteins that lead to Bax and Bak release, causing it to migrate from the cytosol to the mitochondria, and with subsequent induction of caspase activity to drive cell death. 14
The objective of this study was to identify whether the regulation of PUMA by the combined use of hypoxia or doxorubicin decreases the apoptotic process through the activity of hypoxia-inducible factor HIF-1α and the generation of reactive oxygen species generated by said factors in the HT29 colon cancer cell line.
Materials and methods
Biological systems
The HT29 (ATCC® HTB38™) colorectal adenocarcinoma cell line, established from a primary human colon tumor with epithelial morphology and monolayer growth, was used for this study.
Cell culture
The cells were cultured at 37oC, 21% O2 and 5% CO2 in DMEM medium, supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Hypoxia conditions were chemically induced using 100μM cobalt chloride (CoCl2). Cells were treated with 100 μM CoCl2 or 5μM doxorubicin. The stimuli were applied for periods of 0, 12 and 24 hours.
Cell viability
Cells were subjected to the abovementioned conditions and the trypan blue exclusion method was used to determine their viability after the treatment. A 1:5 dilution of the cell suspension was prepared in trypan blue solution and mixed by pipetting. An aliquot of the mixture was then taken and placed in a Neubauer chamber to count viable (uncolored) and non-viable (blue-colored) cells, thus calculating the percentage of viable cells.
Determination of reactive oxygen species (ROS)
To determine the effect of different oxidative stress conditions on the cellular production of ROS, the 5(6)-carboxy-2,7-difluorodihydrofluorescein diacetate (H2DFFDA) fluorogenic marker (Invitrogen) was used. 12 The cells were cultivated in 96-well cell culture plates (Corning Costar®) and their growth was allowed for 24 hours, after this time treatments were carried out according to the condition to be studied: chemical hypoxia or doxorubicin. Then, they were rinsed with PBS 1x.
Cells were incubated at 37°C with the probe at a concentration of 10μM for 30 minutes and rinsed with PBS 1x. Subsequently, they were lysed with lysis buffer and 200μM of the cell lysate were transferred to a 96-well box for fluorescence reading at an excitation wavelength of 492nm and emission wavelength of 527nm, performed in the Cytation 3 Cell Imaging Multi-Mode Reader Imagine Reader (BioTek®). The fluorescence results obtained were normalized by microgram (μg) of protein using the bicinchoninic acid assay (BCA) method with the Pierce™ BCA Protein Assay Thermo Scientific Kit for quantification. 25uL of the protein extract or the blank were added to 200 μL of working solution (50 parts of reagent and 1 part of reagent B) and incubated for 30 minutes at 37°C in a 96-well plate. The absorbance reading on the Cytation 3 Imagine Reader (BioTek®) was taken at an absorbance of 562nm. Protein concentration was determined using a calibration curve constructed with known concentrations of bovine serum albumin.
Activity of hypoxia-inducible factor HIF-1α
Nuclear proteins were extracted using a nuclear extraction kit (Active Motif) according to the manufacturer's recommendations. The cell suspension was centrifuged at 2 000 rpm at 4°C and the pellet was resuspended by pipetting in hypotonic buffer and transferred to a 1.5mL eppendorf tube, where the vortex suspension was mixed and incubated for 25 minutes on ice. The pellet was resuspended in lysis buffer (supplemented with Ditiotreitol [DTT] at a final concentration of 1mM) and an additional 2.5μL of detergent was added. This suspension was incubated for 30 minutes on a shaker platform at 150 rpm, after which it was centrifuged at 14 000 rpm for 10 minutes.
HIF-1 activity was evaluated from 20μg nuclear extract protein with the TransAM™ HIF-1 Transcription Factor Assay kit. To this end, the 20μg nuclear extract were diluted in lysis buffer (Trans AM™ lysis buffer, with 1mM DTT and protease inhibitor cocktail) up to a volume of 10μL. 40μL of binding buffer (AM4 Buffer and 1mM DTT) and 10μL of nuclear extract were added to wells previously coated with oligonucleotide containing the hypoxia responsive element (HRE).
This mixture was incubated for 1 hour under agitation at 100 rpm. Then three washes were performed with 1x wash buffer and anti-HIF1 antibody (1:500 dilution) was added and incubated for 1 hour in agitation. Incubation was performed with HRP (horseradish peroxidase) secondary antibody conjugate (dilution 1:1000) for 1 hour, after which washes were performed, and the wells were incubated with the dye solution for 10 minutes, then the reaction stopping solution was added. The absorbance reading was made at 450nm with a reference wavelength of 655 nm160.
Caspase-3 Activity
The caspase-3 fluorimetric determination kit (Invitrogen) was used to evaluate apoptosis under different hypoxic conditions. The cells were incubated briefly at a confluence of 80% according to defined treatment times and untreated cells were used as negative control. Then, the medium was aspirated, the culture box was placed on ice and 200μL of lysis buffer (4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (HEPES) pH 7.4 200 mM, 3-[(3-Colamidopropyl)- dimethylammonium]-propane sulfonate (CHAPS) 25 mM and dithiotreitol (DTT) 25 mM) were incubated with this buffer for 10 minutes.
Subsequently, the adhered cells were scraped off and transferred to an eppendorf tube where they were centrifuged at 13 000 rpm for 15 min at 4°C, after which the supernatant was collected and the protein quantified by the BCA method. 20μg protein was mixed with 2.5μL Ac-DEVD-AMC (Acetyl-Asp-Glu-Val-Asp-7-amido-4-methylcoumarin) substrate in a final concentration of 20μM and assay buffer (HEPES 200 mM, pH 7.4 with 1% CHAPS, 50 mM DTT, 20 mM EDTA) to a final volume of 250μL. From this mixture, 200μL were transferred to a 96-well plate for fluorescence reading (Xexcitation: 380nm /λemission: 460nm). Untreated cells were used as negative controls and cells treated with staurosporine (1 ug/ ml) for 4 hours as positive controls of apoptosis. The results were expressed as times of change in fluorescence levels per μg of protein with respect to untreated cells.
mRNA expression of pro-apoptotic genes PUMA and BAX by RT-PCR
RNA extraction was performed after the treatments for each condition to be evaluated were completed. For this purpose, 1mL of Trizol® (Life Technologies) was added, removing the cells adhered by resuspension with this agent, and the suspension obtained was transferred to a 1.5mL eppendorf tube free of ribonucleases. 200μL of chloroform were added with inversion agitation and incubation for 3 minutes at room temperature, after which the mixture was centrifuged at 12 000g at 4°C for 15 minutes (Sigma Centrifuge 1-15K). The supernatant (aqueous phase) was transferred to a 1.5mL ribonuclease free eppendorf where RNA precipitation was performed with 500μL of cold isopropanol and incubation for 10 minutes. The mixture was centrifuged at 12 000g for 10 minutes at 4°C; the supernatant was discarded and the pellet was washed with 75% ribonuclease-free ethanol, and then centrifuged at 12 000g for 5 minutes at 4°C.
The supernatant was discarded and the pellet was resuspended in 50μL of molecular biology quality water (95284 Sigma-Aldrich®). RNA was quantified in the NanoDrop 2000 spectrophotometer (ThermoFisher Scientific®) and the 260/280 absorbance ratio was used to analyze the quality of the extracted RNA. Reverse transcriptase (Maxima First Strand cDNA Synthesis Kit for RT-qPCR - Thermo Scientific) was used for the synthesis of cDNA. For each cDNA synthesis reaction, 4μL of 5x reaction mix, 2μL of maxima enzyme mix, 11 μL of RNA of 500 ng/μL concentration treated with DNAse I and 3μL of nuclease-free water were mixed to a final volume of 20μL for 15 minutes at 50°C, ending the reaction at 85°C for 5 minutes.
For the quantitative evaluation of the changes in mRNA levels of the PUMA and BAX genes, the qRT-PCR technique was used. Specific primers were used for each of these genes (Table 1) and the Platinum®SYBR® Green qPCR SuperMix-UDG cocktail was used, which comes in a 2x concentration and contains Platinum® Taq DNA polymerase, SYBR® Green I Tris-HCl dye, KCl, 6 mM MgCl2, 400μM dGTP (deoxyguanosine triphosphate), 400μM dATP (deoxiadenosine triphosphate), 400uM dCTP (deoxycitidine triphosphate), 800μM dUTP (deoxiuridine triphosphate), uracil-DNA glycosylase and stabilizers. For each gene to be evaluated, a reaction mixture was prepared and the specific qRT-PCR program was run in the DNA Engine Opticon® 2 System detection system.
Inhibition of transcription factor HIF-1α
The effect of APE-1 E3330 endonuclease inhibitor on HIF-1α activity was evaluated. The cells treated with the inhibitor were used to evaluate the amount of ROS and the activity of HIF-1α. The cells were incubated with E3330 (10-30 μmol/L) in the presence or not of CoCl2, previously described for the different study conditions.
Results
CoCl2-induced hypoxia generates a variation in the number of reactive oxygen species
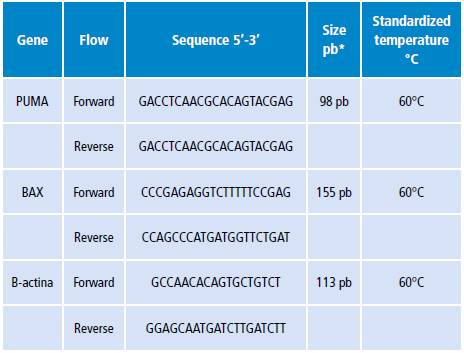
HT29 cells were treated with CoCl2 for 12 and 24 hours to induce HIF-1α activity. CoCl2 100uM increased 14 times the activity of HIF-1α at 24 hours compared to the control group (cells under normoxic conditions) with a p<0.05 (Figure 1). The combined treatment of CoCl2 with doxorubicin showed that HIF-1α activity does not vary with respect to the control with a p>0.05 (Figure 1).

Source: Own elaboration
Figure 1 HIF1-α activity. HT29 cells treated with 100μM CoCl2 or 5μM doxorubicin (CoCl2 + Doxo).
The E3330 inhibitor was used to demonstrate that HIF-1α activity depends on the oxidative stress generated by CoCl2 and doxorubicin; it acts on the APE-1 endonuclease, has a redox regulatory activity and also modulates AP-1 transcription factor activity. 14 Thus, the use of E3330 inhibitor decreased HIF-1α activity modulated by CoCl2 or the use of doxorubicin at around 24 hours compared to control (untreated) cells, p<0.05 (Figure 2).
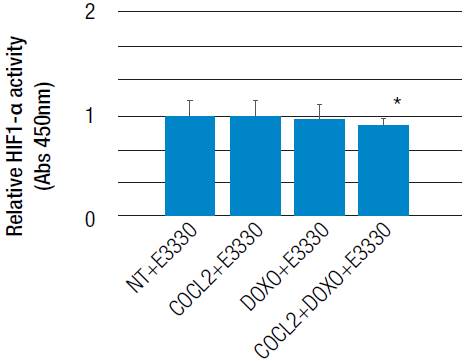
The modulation of oxidative stress could be involved in hypoxia-inducible factor activity
Treatment with CoCl2 and doxorubicin separately generated an increase in reactive oxygen species at 12 and 24 hours. However, it is evident that the treatment with doxorubicin significantly increases the accumulation ofreactive oxygen species compared with the control and with the use of CoCl2. Combined treatment of CoCl2 and doxorubicin has shown a significant increase in the amount of ROS and is similar at 12 and 24 hours (p<0.05 in both cases; Figure 3). Taking into account the previous result of HIF-1α activity due to the effect of CoCl2 and doxorubicin, it could be said that the accumulation of ROS induced by the stimulation with these two agents may be more dependent on the use of doxorubicin than on HIF-1α activity generated by CoCl2. This result may indicate a regulation of the transcription factor HIF-1α due to the accumulation of ROS generated by the use of CoCl2 and doxorubicin.
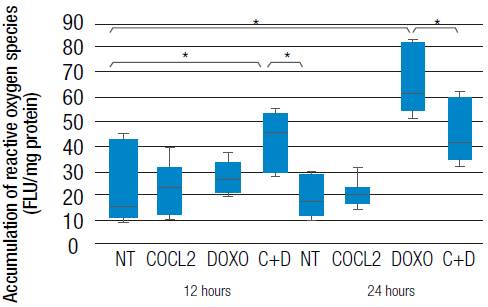
Hypoxia-inducible factor HIF-1α does not alter cell viability in HT29 colon cancer line
It has been considered that an increase in the amount of reactive oxygen species could lead to cell death. For this reason, HT29 cells viability under chemical hypoxia conditions (100uM CoCl2) and the combination of treatments (l00uM CoC12 + 0.5 μM doxorubicin) (Figure 4) at different times (0-24h) was evaluated. The results show that there is no significant variation (p>0.05) in cell viability at different times and as a result of the aforementioned stimuli. Likewise, the treatment of HT29 cells with the combination of CoC12 and doxorubicin treatment maintained cell viability at different times (0-24h), compared with untreated cells, with p>0.05 (Figure 4) and positive control H2O2 (200uM)p>0.05.
The process of apoptosis may be controlled by hypoxia
After 6 hours of incubation with CoCl2 (100uM) under the conditions described above, caspase-3 activity decreased, which is attributed to increased HIF-1α activity over time. Caspase-3 activity decreased after 6 hours (p<0.05 relative to times of l and 3 hours) (Figure 5).
Figure 5 also shows how the effect of the combination of CoCl2 with doxorubicin decreases caspase-3 activity (p>0.05) compared with normal cells between hours 1 and 12. However, caspase-3 activity increases at 24 hours. It could be considered that the effec of these stimuli on caspase-3 activity is time dependent and could be caused by HIF-1α activity.
The expression of PUMA and BAX pro-apoptotic genes could be modulated by HIF-1α and ROS
The qRT-PCR technique allowed finding that treatment with l00uM CoCl2 significantly decreased PUMA mRNA expression (p<0.05): in turn, the combined treatment of CoCl2 or 5uM doxorubicin led to a significant increase (p<0.05) in PUMA mRNA expression, whik BAX mRNA expression was significantly reduced (p<0.05) (Figure 6). The E3330 inhibitor led to a decrease in mRNA expression of the pro-apoptotic PUMA gene. This decrease is around 10 times lower with a p<0.05 compared with cells without treatment (Figure 7).

Source: Own elaboration
Figure 6 Relative PUMA and BAX mRNA expression. HT29 cells were treated with 100 μM CoCl2 or 5 μM doxorubicin (CoCl2 + Doxo) for 24 hours
The expression of the PUMA pro-apoptotic gene could be modulated by the accumulation of ROS
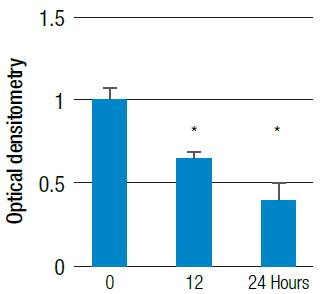
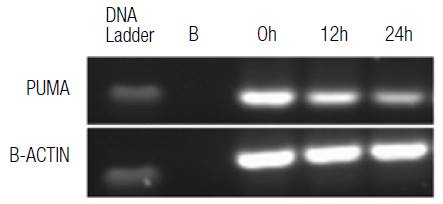
One of the possible mechanisms by which oxidative stress can induce cell death is apoptosis; for this reason, PUMA mRNA levels were evaluated through conventional PCR. Hydrogen peroxide is a reactive species modulator that was used at a concentration of 200uM to determine the effect on the alteration of PUMA mRNA expression. This effect led to a decrease in PUMA mRNA expression (p<0.05) with respect to untreated cells (Figure 8 and 9). These results could indicate that the increase in the generation of ROS by hydrogen peroxide is comparable with the use of HIF-1α and doxorubicin separately, which significantly increases the amount of ROS and, in turn, significantly reduces the expression of PUMA mRNA.
Source: Own elaboration
Figure 8 Relative PUMA mRNA expression. HT29 cells were treated with 200 μM H2O2 at different times, evaluated by conventional PCR for PUMA and compared to the b-actin gene expression in agarose gel.
Discussion
Resistance in cancer can be measured through different cellular mechanisms, including apoptosis. 15 A hypoxic tumor environment is considered as one of the critical factors that favor drug resistance through the MDR phenotype. 16 Likewise, it is known that there is a wide range of molecules that could be regulated by hypoxia, with EHF-1α being the main response factor that modulates several processes. 17
A hypoxic environment can be comparable to the effect produced by the use of substances such as CoCl2, which acts as an iron chelator necessary for prolyl-hydroxylases to act by hydroxylating HIF-1α and leading to the translocation of EHF-1α to the nucleus and its dimerization with EHF-1β, forming EHF-1. On the other hand, doxorubicin exerts antitumor activity inhibiting the proliferation of different cancer cell types 18,19; its use has been limited by an increase of oxidative stress and because it is a transporter substrate associated with drug resistance such as Pgp. However, its action, considering the hypoxic tumor environment, has not yet been well documented.
Previous studies have shown that both HIF-1 and HIF-2 could modulate the response to Sunitinib treatment in colon cancer cells leading to a decrease in cell proliferation. 20,21 Therefore, it is important to consider hypoxia inducible factor-1alpha activity in different molecular events necessary to regulate tumorigenicity. The transcription factor HIF-1α may be associated with the control of cell death by regulating the expression of pro-apoptotic genes.
The correlation between HIF-1α and the PUMA and BAX genes as pro-apoptotic inducers is not clear yet. This study demonstrated that chemical hypoxia induced with CoCl2 promotes a decrease in the expression of PUMA mRNA level and maintains the basal expression of BAX mRNA. The same situation was observed in the determination of the effector caspase-3 activity in the apoptotic process. On the other hand, using doxorubicin alone caused a significant decrease in the expression of PUMA and BAX mRNA. However, when treatment with CoCl2 was combined with doxorubicin, the PUMA mRNA expression increased, but HIF-1α activity decreased, corroborating the correlation between this transcription factor and the PUMA gene.
It should be noted that many solid cancers are highly hypoxic, which would indicate that, in these zones, the expression of the transcription factor HIF-1α is critical for tumorigenicity, favoring a greater proliferation and decreasing the capacity of the apoptotic pathway. Therefore, hypoxia could mediate cell survival by reducing the expression of pro-apoptotic genes, increasing drug resistance of colon cancer cells.
Different studies have shown that redox state altered by hypoxia, anoxia, oxidative stress and generation of ROS could regulate the expression of the PUMA gene in vitro and in vivo. 22-24 Although ROS are generated during apoptosis mediated by oxidative stress, as a result of mitochondrial damage 25, PUMA induction by ROS could induce an upstream feeding mechanism that favors the apoptotic process.
In addition, it has been proven that, in a hypoxic tumor environment, the amount of ROS is high and can activate hypoxia-inducible factors 26, indicating a direct correlation between these factors and oxidative stress. This study showed that the greatest amount of reactive species was observed at 24 hours, when HIF-1α activity is much higher in the HT29 colon cancer cell line, using chemical hypoxia HIF-1α. On the other hand, resistance associated with cell death could be evidenced in this study, where the combination of HIF-1α + doxorubicin promoted the maintenance of cell survival by decreasing the expression of pro-apoptotic factors and keeping cells alive, in such a way that the action of the drug was attenuated.
It has been reported that HIF-1 and the p53 tumor suppressor protein are involved in the cellular response to hypoxia; however, the way these two transcription factors determine cell fate is still unknown. 27 PUMA may activate by being dependent on p53 or not, which could explain the increase when the combination of hypoxia and doxorubicin occurs at 24 hours.
The PUMA protein is a general sensor of cell death stimulation and a promising therapeutic target in cancer 27; therefore knowing the mechanisms associated with its regulation is of great importance. This is also a mediator of p53-dependent apoptosis in a large number of cell types. 28 For example, HCT116 PUMA-knockout colon cancer cells are highly resistant to apoptosis induced by overexpression of p53 or DNA-altering agents such as doxorubicin, 5-fluorouracil (5-FU), cisplatin, oxaliplatin, etoposide and camptothecin. 24,25
Different cell lines have shown that, in response to DNA damage, p53 is activated and leads to an increase in the expression of pro-apoptotic genes such as Noxa, PUMA and Fas. Similarly, cancer cells resistant to cisplatin have shown that there is an alteration of the mechanisms mediated by p53 inducing a decrease in the pro-apoptotic genes Noxa and PUMA. 29
This study showed that there is a decrease in PUMA mRNA expression associated with hypoxia, which in turn is maintained when doxorubicin is used in combination with chemical hypoxia. Therefore, it is possible to consider that there is a modulating effect of HIF-1α on p53 that would affect PUMA expression by altering the apoptotic process of HT29 colon cancer cells, even in the presence of doxorubicin, a drug aimed at DNA alteration. In addition, this research allowed demonstrating that both chemical hypoxia and the use of doxorubicin increase oxidative stress.
APE1 endonuclease (Human AP Endonuclease) participates in redox regulation influenced by stress response, DNA repair and other cellular functions such as angiogenesis, inflammation and cell survival. 30 However, some recent studies show that there is still a wide arrange of unknown activities of this factor. 31 Some APE1 redox signaling targets have been considered, namely p53, AP-1 and HIF-1α, which lead to establish that APE1 inhibition could be considered as therapeutic targets with important clinical potential.32
In this study, APE1 inhibition led to an even greater decrease in the PUMA mRNA expression level, which would confirm the ability of this endonuclease to directly or indirectly regulate the expression of this gene. It is important to consider that HIF-1α regulates the expression of APE1 levels and this, in turn, is regulated by APE1. 33 On the other hand, it is also important to consider that the expression of the pro-apoptotic protein PUMA is regulated by other mechanisms independent of p53, such as induction mediated by the transcription factors Spl, E2Fl and FOXO3a; the latter involves the inhibition of the Akt survival pathway and the activation of the JNK pathway. 33-36 For this reason, factor HIF-1α inhibition does not a guarantee the activity of a drug, and it is necessary to take into account its capacity, in a hypoxic environment, to increase the reactive oxygen species and the alteration of factors that regulate the cellular redox state that may occur.
Conclusion
This study demonstrates that hypoxia could lead to maintain cell survival of the HT29 colon cancer line by keeping HIF-1α active. Hypoxia-inducible factor activity can be favored by the increase of reactive oxygen species and, thus, modulate the expression of the pro-apoptotic factor PUMA. However, the inhibition of this transcription factor does not guarantee an increase in the expression of PUMA mRNA, since this could be dependent on the generation of ROS and, in turn, the redox regulatory factor APE1. This factor may have an important role in the regulation of the transcription of pro-apoptotic genes. The results of this study suggest that the inhibition of factors such as hypoxia or ROS could be useful, as a therapeutic target, to enhance the effectiveness of drugs used in cancer.