Introduction
The potato (Solanum tuberosum L.) is crucial for global food security and nutrition, after rice, wheat, and maize (FAOSTAT, 2021). Due to population growth, increased food demand, pest and disease pressure, and climate change, there is a need to improve potato production by developing new varieties that use various genetic resources, including species of wild and cultivated Solanum (Stokstad, 2019). However, many cultivated potato varieties still exhibit a narrow genetic base. This limitation can be attributed to decades of repeated breeding aimed at improving specific agronomic characteristics (Ruiz-de Arcaute, 2022).
Initially, the characterization and evaluation of potatoes was based on morphological and physiological traits, including plant architecture, tuber shape, skin color, and flesh color. Today, breeding programs emphasize the use of advanced genetic and molecular techniques to develop superior potato varieties with improved characteristics and yield. The characterization of wild potato species is essential before implementing them in breeding programs. Molecular markers, particularly simple sequence repeats (SSR), are widely used to study closely related genotypes due to their codominance, locus specificity, reproducibility, and high-throughput genotyping capabilities (Bhardwaj et al., 2023; Ghislain et al., 2009). Additionally, knowing the ploidy of a species could be of great help for taxonomic characterization in breeding programs (Ordoñez et al., 2016). In the specific case of the potato, the basic chromosome number is 12, and although it ranges from diploid to hexaploid, most species are considered diploid (Watanabe, 2015).
Peru, the main center of origin of the potato, shows great biodiversity as a result of an Andean culture that has conserved its resources for generations and that nowadays constitutes an important legacy for humanity (Stokstad, 2019). Within this diversity of potatoes, numerous shapes, colors, textures, and flavors have been distinguished and appreciated for centuries by rural families and, currently, also by urban dwellers, which has increased the commercial demand (Fonseca et al., 2014; Tobin et al., 2018).
22 % of the cultivated area for potato in Peru, distributed in 19 of its 26 regions, corresponds to native potatoes (MIDAGRI, 2017). However, this diversity has not yet been characterized by regions, much less by communities. Each community has different species and varieties of native potatoes that are not yet part of collections or germplasm banks, nor is the degree of “genetic similarity” between them known (De la Cruz et al., 2020). In the Pasco region, located in the Peruvian central Andes, native potatoes are cultivated at more than 3500 masl, whose diversity could be very useful for a genetic improvement program. The Daniel Alcides Carrión
National University (UNDAC) has a project whose objective is to build a germplasm bank of native potatoes representative of the region. From an initial collection of 358 landraces from the Pasco region, this study aims to evaluate and characterize 40 landraces with morphological descriptors, SSR markers, and ploidy levels.
Materials and methods
Plant material
Based on market performance and seed availability, 40 landraces were selected from the native potato collection made by the UNDAC Native Potato Project in the Pasco region in 2019, representing the districts of Paucartambo, Yanahuanca, and Santa Ana de Tusi (Table 1). These landraces were initially identified by folk names in Quechua and Spanish languages, assigned by the local farmers according to their perception about the tuber’s shape, color, texture and other aspects (Domínguez, 2020). The selection was installed in the greenhouse of the School of Agronomy of the Paucartambo branch of the UNDAC (10°46’14.1”S, 75°48’53.4”W, 3200 masl). There were 4 repetitions per landrace, planted individually in 7-liter pots, and arranged in rows with at a distance of 0.9 m x 0.4 m.
Morphological characterization
The first description of the selected landraces was made in the greenhouse when the potato plants were in full bloom and after harvest, following Gómez (2000). The 20 qualitative descriptors considered are detailed in Table 2.
Molecular characterization
For this part, samples composed of leaflets of each landrace (to obtain samples of ca. 100 mg of dry weight) were taken to the Research and Development Laboratory, Genomics Unit of the Cayetano Heredia University in Lima, to perform the following procedures:
DNA extraction: It was performed using the modified CTAB method (Doyle and Doyle, 1987), for which leaflets were used. The DNA quality and quantity were verified by spectrophotometry using an Eppendorf BioPhotometer, and by electrophoresis on 1 % (w/v) agarose gel. The DNA samples were diluted in nuclease-free water at a concentration of 40 ng/ul.
Amplification of microsatellite markers: Amplification reactions were performed with the microsatellite markers used by Ghislain et al. (2006), 2009) (Table 3). The composition of the PCR reactions, the volumes and final concentrations of the reagents were as follows: 1X colorless GoTaq Flexi Buffer, 2.5 mM magnesium chloride (MgCl2), 0.2 mM deoxynucleotide triphosphates (dNTPs), 0.3 uM of each primer, and GoTaq G2 high-throughput DNA polymerase at 5 U/ul. The amplification conditions consisted of an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of denaturation amplification at 94 °C for 1 min, 55 °C for 30 s, and 72 °C for 1 min, followed by a final extension step at 72 °C for 10 min. Amplifications were performed in an Eppendorf Mastercycler.
Following the recommendation of the LI-COR Biosciences manual for microsatellite analysis (LI- COR, 2003), the amplification products were diluted in a Blue Stop Solution 4300 (1:1), denatured for 5 min at 94 °C, and the samples were snap chilled on ice; a 1 μl sample was loaded into each well of a polyacrylamide gel composed of 20 ml of KB Plus Gel Matrix 6.5 %, 150 μl of 10 % ammonium persulfate, and 15 μl of tetramethylethylenediamine (TEMED) with the Tris Borate EDTA Buffer. The pre-run was programmed at 1500 V and 40 W for every 20 min to focus the laser at 700 and 800 nm. The SSR DNA fragments were visualized through 700 or 800 infrared fluorophore dyes after running for 1.5 h at 45 °C, 1275.1 V, and 39.8 W. The molecular weight marker 50 - 350 bp IRDye (700 or 800) was used.
The images obtained were calibrated with the software SagaGT TM version 3.3, following the instructions of the above-mentioned manual. This program allowed visualizing and calibrating the molecular weights for the bands obtained, which were recognized by lasers at wavelengths of 700 nm. The gel calibration was performed using LI-COR molecular weight markers, and the characterization was performed manually using the SagaGT program, considering only sharp bands. Finally, the information of the molecular weights of all the alleles was also included in Table 3.
Data analysis
Genetic diversity: The data analysis was carried out using the R statistical language packages (R Core Team, 2021) Adegenet (Jombart, 2008) and Polystat (Clark, 2011), with which the values of diversity (observed and expected heterozygosity) and number of alleles were found. The distance matrix was determined using the Jaccard similarity index.
The polymorphism information content (PIC) for each SSR locus was calculated as follows:
Where fi is the relative frequency of the i th allele of the SSR loci (Nei, 1973)
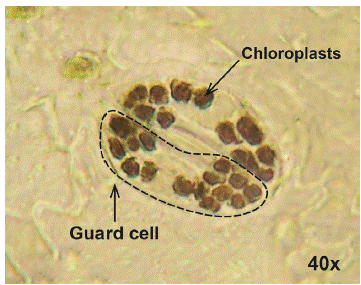
Ploidy level: The ploidy level was determined indirectly (Huamán, 1995) with the technique of counting chloroplasts in guard cells from the epidermis of terminal leaflets of potato plants. The young potato leaf was soaked in 96 % alcohol for 20 min, then the epidermal tissue was removed from the leaf underside with fine forceps, and later it was submerged in a Lugol solution for 2 min. The sample was placed on a slide and covered with a coverslip, after adding a drop of immersion oil for a better visualization of the samples. Finally, the chloroplasts were counted twice with an optical microscope with a 40X objective.
Statistical analysis
The information obtained was stored in a database distinguishing qualitative and quantitative variables. The data was processed in the InfoStat software version 2020. Cluster analysis was performed to generate a similarity dendrogram for qualitative and quantitative variables, based on Ward’s aggregation criterion.
Results
Grouping potato landraces based on morphology
The 40 native potato landraces from the Pasco region were grouped based on 20 morphological descriptors (Figure 1) at a similarity coefficient distance of 9.5, resulting five groups. The first group (A) was made up of 2 subgroups with 5 landraces in total; the second one (B) presented five landraces with 15 % similarity in the form of tuber (elongated), and tuber skin color (yellow); and the third one (C) presented 10 landraces with 20 % similarity, predominating the flower and tuber pulp color, purple and creamy yellow, respectively. The fourth group (D) had eight landraces with 15 % similarity, whose main characteristics were predominantly purple and lilac flower color, oblong tuber shape, and blackish and white skin and pulp color, respectively. Finally, the fifth group (E) was made up of 12 landraces with 20 % similarity, whose main characteristic was the lilac flower color. The least related landraces were Yawar papa, Suytu amarillo, Negra andina and Azul Huayro, located at the extremes of the dendrogram (Figure 1). Likewise, Yana runtush and Azul huayro belonged to the same group, but were distant probably due to the high diversity and polymorphism between them.
Genetic diversity in the potato landraces
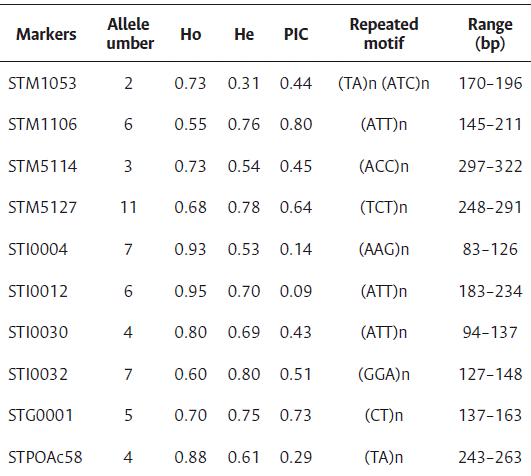
The highest and lowest number of alleles were found for the microsatellites STM5127 (11) and STM1053 (2), respectively (Table 3). The average number of alleles was 5.5, lower than those reported in the literature, 6.8 by Rocha et al. (2010), and over 4 by Tiwari et al. (2019). However, of the total assessed landraces (40), two were excluded from the analysis due to the high number of null alleles generated.
The highest and lowest expected heterozygosity values were expressed by the markers STI0032 (0.795) and STM1053 (0.313), respectively (Table 3). It is considered that the most informative markers were those that presented a positive value when subtracting the expected heterozygosity from the observed one, being the following: STG001, STM1106, ST0032 and STM5127. The lowest PIC value (0.14) was obtained with the marker STI0004, and the highest PIC value (0.80) was obtained with the marker STM1106.
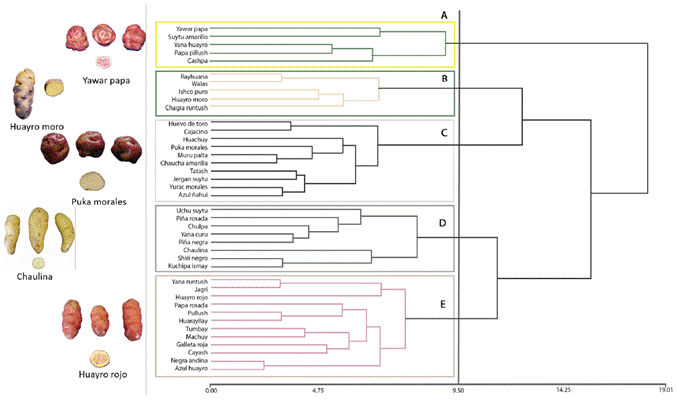
To analyze the genetic relationships between the 38 suitable landraces, the binary matrix of registered alleles was considered and the binary genetic distance between all pairs of genotypes was calculated, taking into account the complete set of markers, mono and polymorphic. The binary genetic distances observed were used to visualize the groups of similar genotypes, by obtaining a dendrogram elaborated through the UPGMA algorithm. Figure 2 shows the high genetic similarity between the landraces Yurac morales and Puka morales; Azul huayro, Yana curu, Tatash, Machuy, and Galleta roja; and with less similarity, Uchu suytu and Cayash. In addition, unique landraces can be seen in the dendrogram: Ishco puro, Negra andina, Yana runtush, and Piña rosada.
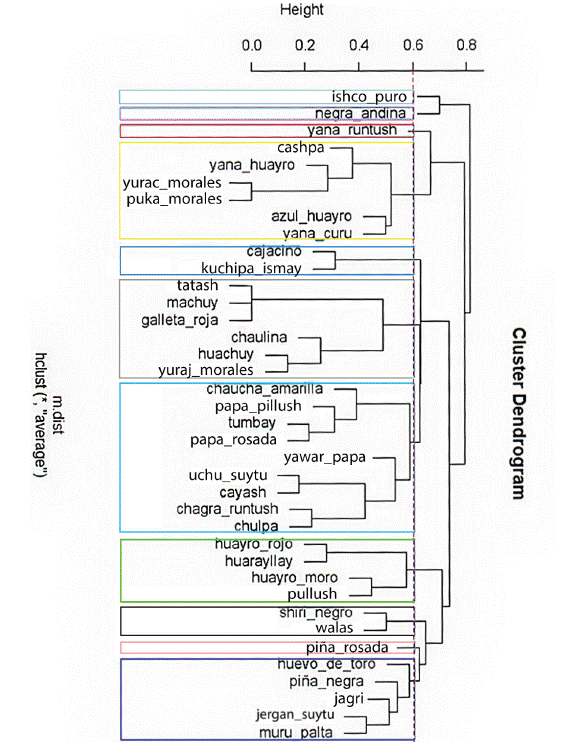
Figure 2 Dendrogram of 40 native potato landraces of the Pasco region considering genetic distance within a molecular characterization and using the UPGMA method (Ward’s aggregation criterion).
Seven groups of native potatoes related at molecular level were identified. The first formed by Cashpa, Yana huayro, Yurac morales, Puka morales, Azul huayro, and Yana curu. In the second group, Cajacino and Kuchipa ismay were observed. In the third group, six landraces were observed: Tatash, Machuy, Galleta roja, Chaulina, Huachuy, and Suytu amarillo. In the fourth group were Chaucha amarilla, Papa pillush, Tumbay, Papa rosada, Yawar papa, Uchu suytu, Cayash, Chagra runtush, and Chulpa. The fifth group was formed by Huayro rojo, Huarayllay, Huayro moro, and Pullush. In the sixth group were Shiri negro and Walas; and in the seventh group, Huevo de toro, Piña negra, Jagri, Jergan suytu, and Muru palta.
3 groups were formed based on the chloroplast count technique (Figures 3 and 4). The first group was made up of the landraces Chaulina, Tumbay, Azul ñahui, Suytu Amarillo, and Ishco puro, which presented eight chloroplasts, and are considered diploid (2n = 2x = 24). In the second group, made up of the landraces Papa pillush, Huachuy, Cajacino, Yana huayro, Negra andina, Chulpa, Piña rosada, Chagra runtush, Chaucha amarilla, Huayro moro, Huayro rojo, Jagri, Shiri negro, and Walas, the number of chloroplasts varied from 9 to 11, corresponding to triploids (2n = 3x = 36). The third group was made up of the landraces Pullush, Huarayllay, Papa rosada, Cashpa, Muru palta, Tatash, Puka morales, Yurac morales, Kuchipa ismay, Yana runtush, Rayhuana, Azul huayro, Piña negra, Uchu suytu, Cayash, Galleta roja, Huevo de toro, Machuy, Yana curu, Yawar papa, and Jergan suytu, ranging from 12 to 14 chloroplasts, considered tetraploids (2n = 4x = 48). Within the 40 landraces evaluated, 5 diploid, 14 triploid, and 21 tetraploid landraces were identified, which maintain all ploidy levels (2n = 2x = 24 to 2n = 4x = 48).
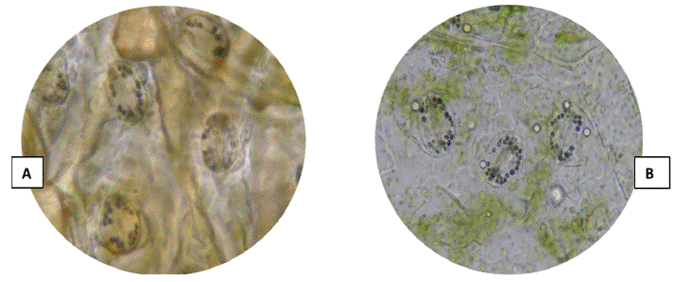
Figure 3 (A) Chloroplasts of the Kuchipa Ismay landrace. (B) Chloroplasts of the Huayro moro landrace.
Discussion
The formed groups of landraces of native potatoes from Pasco, Peru, showed high chromatic diversity both in skin and pulp, unlike what was reported by Calliope (2018) about Andean potato genotypes from Argentina. Monteros et al. (2011) identified a wide variety of primary and secondary skin colors in native potatoes from Peru and Ecuador; however, the main pulp colors in the current research were cream and yellow. Furthermore, the predominant skin color was yellow and blackish, which mostly did not present a secondary skin color, and the predominant flesh colors were white and cream. Since the combined analysis of morphological and molecular markers evaluates different characters, this was not carried out; however, it has been found that they complement each other. Abebe et al. (2013) affirm that the characteristics of potato tubers are subject to interactions between the environment and the genotype.
Native potatoes are characterized by their wide genetic variability and distribution, being a potential to face climate change and guarantee food security. The Pasco region presents a wide diversity that must be conserved and exploited by rescuing ancestral knowledge associated with Andean crops (Rosero et al., 2020; Berdugo et al., 2017). Recently, Zevallos et al. (2021) evaluated the response of native potatoes from this region to Phytophthora infestans, evidencing the potential of the local biodiversity to provide genetic resources for food security.
In this study, ten SSR markers were used to conduct an extensive analysis of genetic diversity and varietal identification among 40 potato landraces. The evaluation of these germplasms offers promising prospects for enhancing sustainable food production in Peru. Moreover, they represent valuable reference materials for the development of hybrid potatoes, serving as crucial parental lines in the breeding programs within Peru. Additionally, molecular markers are considered highly informative when their PIC value exceeds 0.5, moderately informative when the PIC ranges between 0.5 and 0.25, and low informative when the PIC is less than 0.25 (Mahalle, 2023). However, only four SSR markers used in this study (STM1106, STM5127, STI0032 and STG001) were highly informative (PIC>0.5), suggesting their potential use for genetic diversity study on potatoes.
After identifying native potatoes at the morphological and molecular level, the next step would be a study at the functional level, as reported by Martínez et al. (2015) who investigated the functional characterization of starches in Solanum phureja to generate added value, or what was reported by Vargas et al. (2016) in potatoes of Unica variety for specific uses in order to use them in the food industry.
Regarding the genetic relationship between the landraces, the low number of alleles found in this study may be due to the number of microsatellites used and landraces evaluated. Additionally, Onamu et al. (2012) stated that molecular markers probably have a greater influence on the comparison of landraces due to the large number of loci. The chloroplast count technique in the guard cells of the stomata does not accurately determine the ploidy of genotypes, however, it allows the diploid group to be quickly discriminated from other groups (Sánchez, 2017). The number of chloroplasts is related to the ploidy level, which agrees with Huamán (1995). Among the formed groups, the most and least abundant were tetraploids and diploids, respectively, possibly because farmers grow native potatoes in complex mixtures, associated with quality traits such as texture and flavor, according to De Haan (2009).
Conclusions
This study represents the initial morphological and molecular approach to investigate the genetic diversity of potatoes in the Pasco region. It was possible to characterize 40 promising native potato landraces segregated into 5 groups with the first criteria, and 7 groups and 4 unique landraces with the molecular approach. Low diversity was revealed when using a set of 10 SSR markers, indicating limited applicability for studying the genetic diversity of local potato landraces. However, a higher PIC was observed with the markers STG001 and STM1106. The limitations of these findings highlight the necessity for further studies involving a broader range of markers and a more diverse set of genotypes from the Pasco region.