Introduction
Tuberculosis (TB) is a globally distributed infectious bacterial disease with high morbidity and mortality. In 2020, according to the World Health Organization (WHO),1 9.9 million people contracted this infection worldwide. In Colombia, TB represents a major public health problem because, according to the National Institute of Health (INS by its Spanish acronym), 14 338 cases were reported in 2018, which is equivalent to an incidence of 26 cases per 100 000 inhabitants.2
TB is caused by the bacterium Mycobacterium tuberculosis, which mainly affects the lungs (pulmonary TB), but can also affect other organs (extrapulmonary TB).1 Multidrug-resistant TB (MDR-TB) refers to cases in which M. tuberculosis is resistant to the first-line medication, that is rifampicin (RIF) and isoniazid (H), representing a major public health concern and a barrier to disease control in many countries. In this regard, in 2020, the WHO reported that there were 132 222 incident cases of RIF-resistant TB (RR-TB) and MDR-TB in people with a confirmed diagnosis of pulmonary tuberculosis around the world,1 and in Colombia, according to the INS, the average number of MDR-TB cases per year was 110 between 2012 and 2018.3
RIF resistance is an important epidemiological marker for detecting MDR-TB. Resistance to this drug at the molecular level has been attributed to specific alterations, insertions and deletions in the gene encoding the ß subunit of RNA polymerase, known as rpo ß. In addition, it has been established that more than 95% of isolates with RIF resistance have mutations within an 81-bp region (codons 507 to 533) of the rpo ß gene, often referred to as the rifampicin-resistance determinant region (RRDR).4 It should be noted that, as stated by Adikaram et al.,5 the nature and frequency of mutations in the rpo ß gene of RR-TB clinical isolates vary considerably depending on the geographic location.
The Xpert™ MTB/RIF (Xpert) method is a semi-quantitative real-time polymerase chain reaction (PCR) test that simultaneously detects the genetic material of the M. tuberculosis complex and the rpo ß gene mutations associated with RIF resistance in two hours, thus it has revolutionized TB diagnosis.6 This methodology uses molecular probes in 5 overlapping regions of the rpo ß gene that are distributed as follows: probe A: codons from 507 to 511, probe B: codons from 512 to 518, probe C: codons from 518 to 523, probe D: codons from 523 to 529, and probe E: codons from 529 to 533.6 In 2011, through a policy statement, the WHO6 endorsed the use of this test, leading to its increased use to simultaneously detect TB and RIF resistance, which, in turn, has increased detection of drug-resistant TB cases.1
Data on the prevalence of rpo ß gene mutations in resistant isolates detected using the Xpert method are limited because, in Colombia, many laboratories have only recently started employing it. In this sense, the objective of the present study was to describe the distribution and frequency of potential mutations associated with RIF resistance in the rpo ß gene of M. tuberculosis detected in pulmonary and extrapulmonary samples using the Xpert method.
Materials and methods
Type of study
Retrospective study with convenience sampling. A total of 66 samples (pulmonary and extrapulmonary) with positive results for M. tuberculosis and RIF resistance, processed using the Xpert method between January 2011 and July 2019, were analyzed. 43 samples were collected from patients treated at the Hospital Universitario San Vicente Fundación in Medellín, a tertiary care institution and reference center for infectious diseases in the region, and the remaining 23 were specimens sent by external institutions from the Medellín metropolitan area for the exclusive performance of the molecular test (Xpert); most of the latter were smear positive and came from patients with a high suspicion of MDR-TB.
Clinical information collection
From the patients treated at the Hospital Universitario San Vicente Fundación de Medellín (n=43), information on the following variables was obtained after reviewing their medical records: sex, age, serological status for human immunodeficiency virus (HIV), history of TB, previous antituberculosis treatment, and results of mycobacterial culture. In turn, only basic demographic data (age and sex) and information on the type of sample were collected for samples sent by external institutions (n=13).
Microbiological methods
Samples obtained at the Hospital Universitario San Vicente Fundación were prepared for Ziehl-Neelsen staining. The culture method used until 2015 was Ogawa Kudoh, and then it was switched to MGIT (Mycobacterial Growth Indicator Tube) medium. Cultures positive for M. tuberculosis (n=31) were sent to the Laboratorio Departamental de Salud Pública de Antioquia (LDSP by its Spanish acronym - Departmental Public Health Laboratory of Antioquia) to establish their susceptibility to first-line antituberculosis drugs, including RIF; until 2016, this process was performed using the multiple ratio method,7 and since then, the Genotype® MTBDRplus molecular test has been utilized.
Molecular analysis
Pulmonary samples were processed using the Xpert method following the manufacturer's instructions, while extrapulmonary samples were processed according to an institutional protocol described by Peñata et al.8 in a 2016 study.
As mentioned above, the Xpert method uses five molecular probes (A, B, C, D, and E) corresponding to five overlapping regions of the rpo ß gene. The Dx System software of the instrument used to perform the test (GeneXpert) determines a potential RIF resistance in the probe if the molecular beacon scanned in that probe does not bind to the complementary target of the native gene with a difference >4 cycles in the delta cycle (delta C) relative to the other probes.
Moreover, the semiquantitative microbial load data for each of the samples were recorded in four categories depending on the cycle threshold (Ct): very low, low, medium, and high. These categories are provided directly by the software when it outputs the result.
Statistical analysis
Data were analyzed using descriptive statistics, calculating absolute frequencies and percentages for qualitative variables (such as probes with mutations), and means with their respective standard deviation for quantitative variables. All statistical analyses were performed in the statistical software R version 3.6.3.
The different types of study samples disaggregated by semi-quantitative load were presented. Furthermore, concordance (%) between the Xpert method and the tests performed at the LDSP (multiple ratio method and GenoType MTBDRplus® molecular test) to detect susceptibility to first-line drugs (RIF) was calculated.
Ethical considerations
The study was approved by the Institutional Review Board of the Hospital San Vicente Fundación according to Minutes No. 04/2021 of February 12, 2021. In addition, it took into account the ethical principles for research involving human subjects established in the Declaration of Helsinki,9 as well as the provisions on health research set forth in Resolution 8430 of 1993 of the Colombian Ministry of Health.10
Results
The majority of samples were collected from men (63.64%), and the mean age of the patients was 39.60±17.69 years, with a minimum age of 10 years and a maximum of 89 years. A particular finding was that 24.24% of the samples came from patients with HIV infection, although it should be noted that these data was not available for 23 of the 66 patients (34.85%). Other clinical and sociodemographic characteristics are presented in Table 1.
Table 1 Demographic and clinical characteristics of patients with rifampicin-resistant Mycobacterium tuberculosis detected using the Xpert® MTB/RIF method (n=66).
| Characteristics | n | % | |
|---|---|---|---|
| Sex | Male | 42 | 63.64 |
| Female | 24 | 36.36 | |
| Age (years) | 0-18 | 5 | 7.58 |
| 18-24 | 7 | 10.61 | |
| 25-34 | 19 | 28.79 | |
| 35-44 | 10 | 15.15 | |
| 45-55 | 14 | 21.21 | |
| >55 | 11 | 16.67 | |
| Clinical history | HIV: Positive/Negative/ND | 16/27/23 | 24.24/40.91/34.85 |
| Previous TB: Yes/No/ND | 13/30/23 | 19.70/45.45/34.85 | |
| Previous TT: Yes/No/ND | 8/35/23 | 12.12/53.03/34.85 | |
HIV: human immunodeficiency virus; TT: treatment; ND: no data.
Source: Own elaboration.
Regarding the type of sample, it was found that pulmonary samples were the most predominant with 48 cases (72.73%), while most extrapulmonary samples (n=18) were obtained from the lymph nodes (38.88%). The semi-quantitative load of the Xpert method is detailed in Table 2 according to the type of sample, with the "very low" category being the most frequent, with 19 samples (28.78%).
Table 2 Distribution of the semi-quantitative load of the Xpert® MTB/RIF method according to the type of sample.
| Type of samples | n (%) | Semi-quantitative load Mycobacterium Tuberculosis | ||||
|---|---|---|---|---|---|---|
| High | Medium | Low | Very low | |||
| Pulmonary (n=48) | Bronchoalveolar lavage | 33 (50.00) | 7 | 9 | 10 | 7 |
| Sputum | 15 (22.73) | 5 | 4 | 3 | 3 | |
| Extrapulmonary (n=18) | Lymph node | 7 (10.61) | 0 | 2 | 1 | 4 |
| Cerebrospinal fluid | 3 (4.55) | 0 | 0 | 0 | 3 | |
| Pleural fluid | 2 (3.03) | 1 | 1 | 0 | 0 | |
| Vertebral biopsy | 2 (3.03) | 0 | 0 | 2 | 0 | |
| Tracheal aspirate | 1 (1.52) | 0 | 1 | 0 | 0 | |
| Lung biopsy | 1 (1.52) | 0 | 1 | 0 | 0 | |
| Ascitic fluid | 1 (1.52) | 0 | 0 | 0 | 1 | |
| Abscess | 1 (1.52) | 0 | 0 | 0 | 1 | |
| Total | 66 | 66 | 13 | 18 | 16 | |
Source: Own elaboration.
The distribution and frequency of mutations that allowed establishing RIF resistance was 38 (57.57%) for probe E, 16 (24.24%) for probe B, 8 (12.12%) for probe D, 3 (4.55%) for probe A, and 1 (1.5%) for the D&E probe combination. No mutations were detected for probe C (Figure 1).
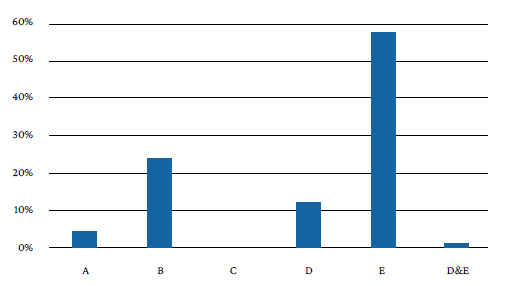
Probes (codons): A (507-511), B (512-518), C (518-523), D (523-529), and E (529-533).
Source: Own elaboration.
Figure 1 General profile of probes with potential mutations in the rpoB gene that confer resistance to Mycobacterium tuberculosis according to the Xpert® MTB/RIF method.
Finally, of the 43 samples cultured for mycobacteria, 8 were negative, 4 were contaminated, and 31 were positive for M. tuberculosis. Of the 31 cultures processed at the LDSP to determine susceptibility to first-line antituberculosis drugs (multiple ratio method and GenoType MTBDRplus® test), it was found that 2 could not be interpreted due to insufficient growth and that RIF resistance was detected in 25, in other words, an 86.20% (25/29) concordance was found with the Xpert method in the detection of this resistance. The probes with mutations associated with the detection of RIF resistance in the 31 positive cultures are outlined in Table 3.
Table 3 Probes with mutations in the Xpert MTB/RIF™ test associated with the detection of rifampicin and isoniazid resistance in cultures positive for Mycobacterium tuberculosis.
| Results reported by the LDSP | Total (n=31) | Associated probes (cases) | |
|---|---|---|---|
| n | % | ||
| MDR | 22 | 71.0 | E (n=19), B (n=3) |
| RIF-R | 3 | 9.7 | D (n=3) |
| H-R | 1 | 3.2 | E (n=1) |
| Sensitive to RIF and H | 3 | 9.7 | E (n=3) |
| Not interpretable | 2 | 6.5 | E (n=1), A (n=1) |
LDSP: Laboratorio Departamental de Salud Pública; RIF: rifampicin; H: isoniazid; MDR: multi-drug resistant (resistance to RIF and H); RIF-R: rifampicin resistance only; H-R: isoniazid resistance only.
Source: Own elaboration.
Discussion
Knowledge of the molecular basis of drug-resistant M. tuberculosis has led to further study the mutations of the different genes involved, which in turn has allowed the development of useful technological platforms for research in this area. The consolidation of molecular tests in Colombia has opened the possibility of strengthening early detection activities and has boosted the development of molecular epidemiology studies on TB.
In the present study, most of the samples positive for M. tuberculosis were collected from men (63.64%), which coincides with data issued by the WHO in its 2021 global report for this disease1 that indicated that in 2020 men accounted for 56% of all TB cases worldwide.
Likewise, in the present study it was found that, of the 43 patients treated at the Hospital Universitario San Vicente Fundación, 37.21% (n=16) had HIV infection, a fairly high percentage compared to that reported by the WHO1 in 2020, which reported that 8% of incident TB cases were living with HIV. In this regard, in a systematic review and meta-analysis, Mesfin et al.11 established that the probability of having MDR-TB was 24% higher among HIV-positive cases. At this point, it should be noted that the Global Laboratory Initiative Technical Report recommends the use of the Xpert method in clinical decision making and follow-up of HIV patients.12
On the other hand, 30.23% of the patients treated at the Hospital Universitario San Vicente Fundación had a history of TB and 18.60% received previous treatment. This is relevant because, as Chonde et al.13 stated in their study of 1 167 patients in Tanzania, resistance to first-line anti-TB drugs in previously treated patients is an indicator of non-adherence and the implementation of an ineffective TB control program.
Although pulmonary specimens (bronchoalveolar lavage and sputum) predominated in the present study (72.73%), the percentage of extrapulmonary samples was quite significant (27.27%). In this regard, it is important to stress that the current WHO recommendations for the use of the Xpert method establish that it should be used as the initial diagnostic test in the analysis of selected extrapulmonary samples (cerebrospinal fluid, lymph nodes, and tissue samples).12
In the present study, rpoB gene mutations were most frequently located in probe E (codons 529-533), with 38 mutations (57.58%), which is in line with the figures reported in the world literature.14-16Furthermore, previous studies carried out in Colombia by Ferro et al.17 and Llerena & Medina18 showed that the most common mutation (36-64% of the total isolates) conferring RIF resistance was located in codon 531 of the rpoB gene, in which serine is replaced by lysine. This mutation has also been reported in several Latin American countries: Zenteno et al.19 reported 47% in Mexico, Asencios et al.20 reported 56.4% in Peru, and Araya et al.2 found 56% in Chile.
Probe B (codons 512-518, associated with Asp516Val) was the second most frequent location of rpoB gene mutations, with 16 mutations (24.2%), which coincides with what was previously reported in Colombia by Llerena & Medina,18 who also ranked this mutation as the second most common, with 21.6%, in their study performed with 689 culture test results.
Mutations in probes D and A were less common and no cases were found in probe C. Previous studies in Nigeria,14 Uganda16 and Ethiopia22 also found no mutations in probe C, suggesting that this location is probably less susceptible to mutations conferring RIF resistance.
In the present study, mutations were more frequent in probes E and B, and less frequent in probes D, A, and C, a finding partially similar to what was described by Reddy & Alvarez-Uria23 in a study conducted in India, in which they identified 171 patients with RIF-resistant M. tuberculosis and reported that mutations in the rpoB gene were indeed less frequent in probes A (8.2%) and C (0.6%), and more frequent in Probes E (94.55%) and B (15.2%). However, it should me mentioned that, with regard to probe D, our findings do differ since the study by Reddy & Alvarez-Uria23 in reports that the second most frequent location of mutations was probe D (18.1%).
Although the results described above reveal that several geographical regions share a high frequency of the same mutations, it is important to keep in mind that these distributions may vary by country. For example, the proportion of mutations in the E and B probes was equal (35.94%) in a study by Chikaonda et al.24 with 64 RIF-resistant M. tuberculosis isolates obtained from 43 adult patients in Malawi. In turn, Alemu et al.,22 in an investigation carried out in Ethiopia with 100 RIF-resistant samples detected with the Xpert method, found that the most common mutation was found in probe E with 81%, followed by probe D with 10%, and probe B with 3%.
In the present study, simultaneous mutations of two probes were found in only one case (D&E), as this is a rare event. Reddy et al.23 found the combination of two probes in five cases (A&D in two cases, A&E in two cases, and B&D in one case), as did Uddin et al.25 (A&E in one case, B&E in one case, A&B in one case, and A&D in two cases). According to the latter authors, who carried out their study on 205 Xpert XR-TB samples detected in Beijing and outside Beijing, simultaneous mutations of two probes may occur in patients with a history of TB, as it has been shown that retreatment of TB increases the proportion of mixed bacilli than primary cases of TB. 25
Regarding concordance by methods performed to cultures to determine susceptibility to RIF at the LDSP, four discrepancies (false resistance by Xpert) were found in the present study. Several causes for this scenario have been reported in the literature, including a heteroresistant bacillary population,19 silent mutations,26 and mutations associated with a low level of RIF resistance undetected using phenotypic methods.27 Moreover, these false resistances were more frequent for probe E (four cases), which coincides with what has been reported in the literature.28,29 Therefore, a positive resistance result in the molecular test should be carefully analyzed taking into account the patient's clinical data; that is, the presence of risk factors in each patient should be evaluated.
The main limitation of the present study was its retrospective nature since, due to the impossibility of retrieving information that may be valuable for a comprehensive evaluation, results from samples sent by external institutions were included, but these samples lacked medical records, mycobacterial culture results, and drug susceptibility tests. Other limitations were the impossibility of sequencing the rpoB gene to identify the specific mutations detected in the Xpert method and to determine the possible explanation for the discordant cases when compared with those analyzed by the LDSP. Nevertheless, this work provides an interesting characterization of RR-TB cases over a long period of time with a technology widely used in the country.















