Introduction
Cacao (Theobroma cacao L.), is a tropical species that provides sustainable economic and environmental benefits to some of the poorest and most ecologically sensitive areas of the world. The upper Amazon, including parts of Ecuador, Peru, Brazil, and Colombia, is generally believed to be the origin center of cacao due to high morphological and genetic diversity observed in this region (Richardson, Whitlock, Meerow & Madriñán, 2015).
In general sense, seeds or grafted seedlings are use as cacao planting materials. Since cacao is naturally cross-pollinated, cacao planting materials from seeds are usually exhibits a highly heterogeneous genetic background. Cacao planting materials propagated clonally through grafting result in both low multiplication rate and undesirable bushy-like growth pattern.
Micropropagation through plant tissue culture expects to overcome these obstacles. Utilizing plant tissue culture technology and molecular biology to accelerate the improvement of cacao breeding programs. In addition, an emergence of molecular marker analyses in genome studies has greatly enhanced the speed and efficacy of cacao breeding programs (Thondaiman, Rajamani, Senthil, Shoba & Joel, 2013).
In fact, in previous breeding studies, numerous populations are sample to detect candidate genes or molecular markers associated with economically important traits. Molecular markers serving as an important tool to check genetic uniformity and nature of micrografted plants (Tiwari, Chandel, Gupta, Gopal, Singh & Bhardwaj, 2013).
Cocoa contains high levels ofpolyphenols, which can interfere with DNA isolation. Furthermore, the presence of inhibitor components can decrease the efficiency or interfere with DNA PCR (Pinto, Forte, Guastadisegni, Martino, Schena & Tantillo, 2007). In this sense, several studies have reported the efficiency of using cocoa leaf material for DNA extraction, however, has not been considered leaf developmental stage as an important variable for obtaining high quantity and quality of DNA.
A modified CTAB method was applied to carry out molecular marker analysis to isolate good-quality DNA from stored leaf tissues (Bhattacharjee, Kolesnikova-Allen, Aikpokpodion, Taiwo & Ingelbrecht, 2004). Alternatively, Ferreira-Santos, Vanderlei, Clément, Pires, Matos, Batista & Peres (2014), used a mixed alkyltrimethylammonium bromide (MATAB) method for high density linkage map using fresh adult cocoa leaves. Saunders, Mischke, Leamy & Hemeida (2004), proved the effectiveness of DNeasy Plant kit® (Qiagen) for DNA extraction to evaluated SSR primers for DNA fingerprinting leaf material. Haymes, Mischke, Scott & Saunders (2004), verified this kit with polyvinyl polypyrrolidone (PVPP) for DNA isolation of cocoa leaf material. Furthermore, DNeasy Plant kit® was used in different studies like genetic stability in cocoa embryo cryoconservation.
Conversely, cocoa genetic identity using molecular markers (Fang, Meinhardt, Mischke, Bellato, Motilal & Zhang, 2014) and epigenetic fidelity of cocoa somatic embryos in cryopreservation and postcryopreservation are reported. However, in the above-mentioned works, leaf developmental stage was not considered as an important variable on DNA extraction considering that it is the most used tissue material and is crucial to obtain high quality and quantity of DNA.
Given these concerns, the aim of this study was to evaluate three different cocoa leaves developmental stages applying some protocols for DNA extraction in field plant (FPL) and in vitro plantlets (IVL). Different PCR amplification conditions were evaluated to obtain a positive amplification for genetic analysis.
Material and methods
Plant material
DNA extraction methods using FPL and IVL regenerated by somatic embryogenesis were evaluated. In this study was used cocoa genotypes TSH565 and CCN51. Plant materials were collected from Nacional de Chocolates Company (CHCN) in Bucaramanga-Santander, Colombia. Cocoa trees were identified and labeled for each selected cocoa genotype from CNCH.
Propagated plant material by grafting in field
To start DNA genomic extraction from TSH565 genotype, three developmental stages of FPL was used. Cacao leaf development proceeds through successive stages categorized as stages A-E (Figure 1a).

Figure 1 Different resources of cocoa leaves, field plants leaves (FPL) and in vitro plantlets leaves (IVL) for extraction of genomic DNA. a) Developmental stages of leaves from field plants of CCN51 genotype. Stage A, emerging cacao leaves with red color. Stage B and C, leaves undergo expansion with reddish to pale brown color. Stage D, leaf accumulates chlorophyll with light green. State E, leaf fully developed with dark green (Mejia, Guiltinan, Shi, Landherr & Maximova, 2012). In vitro plantlets of b) TSH565 genotype and c) CCN51 genotype with 60 days in medium MM6.
Stage A, emerging cacao leaves are translucent and thin, often with distinctive red color and this stage corresponds almost to 10 days during time from leaf emergence. Stages B and C, corresponds to leaves undergo expansion but are still thin, translucent and pigmented and they are 15 and 20 days, respectively. Stage D, is identified 25 days from emergence, where the elongation ceases, and leaf accumulates chlorophyll, becomes light green, leafs non-translucent and begins hardening. Stage E, leaf fully developed, leathery, and dark green with 30 days of development (Mejia et al., 2012). In this study were used C, D, E stages of TSH565 cocoa genotype to evaluate the efficiency of DNA extraction methods.
A total of 5 cocoa trees were labeled for each genotype and through visual inspection were taken on average 10 leaves in good phytosanitary conditions, were cleaned superficially and separated with tissue wipers. Subsequently, were packed into resalable plastic bags and pre-cooled with dry ice to ensure lowest possible temperature during 12 hours of transportation to laboratory. Once in laboratory, they were conserved at -80°C until processing.
Propagated plant material by somatic embryogenesis
In vitro tissue culture experiments were conducted with the same genotypes from CNCH. Immature flowers were stored in sterile basal DKW salts (Ajijah, Hartati, Rubiyo & Sukma, 2016) on ice during transportation and carried out to laboratory. Flower buds were surface sterilized following the protocol described by Urrea, Gallego & Atehortúa (2011), with modifications. Staminodes and petals were extracted from the basal portion of flower bud. SE was induced according to the protocol described by Henao, de la Hoz, Ospina, Atehortúa & Urrea (2018), with some modifications.
DNA Extraction
For genomic DNA extraction of FPL, different methodologies were evaluated, CTAB extraction described by Michiels, Van den, Tucker, Van Riet & Van Laere (2003), and commercial kits such as DNeasy Plant Maxi Kit® Qiagen (Qiagen) and PowerPlant® Pro DNA Isolation Kit MoBio (Qiagen) following the instructions (Table 1).
All protocols have been described as efficient for isolating good-quality DNA from samples with large quantities of secondary compounds. In addition, for removing mucilaginous polysaccharides of cocoa tissues, the different protocols for DNA extraction were carried out with an additional wash with a sorbitol buffer. Finally, once obtained the best method for DNA extraction from FPL, the selected method was used for IVL.
DNA extraction of in vitro plantlets with three months on MM6 medium with leaves in a size between 2-5 cm were selected under sterile conditions using a laminar flow chamber (Figure 1 b-c), leaves were cut and immediately transferred into tubes of 2 ml in liquid nitrogen and taken for storage at -80°C until processing.
Sorbitol buffer was established according to Souza, Muller, Brandao & Lovato (2012), which was added to 0.1 g of leaf tissue, and tubes were placed into cellular disruptor device (BeadBug™), during 5 minutes at 3000 rpm, and subsequently, heated at 65°C. Finally, samples were centrifuged during 10 minutes at 5000 rpm and supernatant was lastly discard. This procedure was repeated twice and continue with the other protocol steps.
DNA Concentration and Purity
The quality and quantity of DNA obtained were determined using microvolume 2000 NanoDrop™ (Thermo Scientific) spectrophotometer and later confirmed on agarose gel 0.8%-stained with ethidium bromide (0.5 mg.mL-1), subsequently visualized under an UV lamp. DNA quantity and purity were evaluated by measuring A260/A280 and A260/A230 absorbance ratios.
PCR amplification Microsatellite Markers
The set of oligonucleotide primers were chosen for presenting the greatest polymorphism mTc-CIR17, mTc-UniCAMP5, mTcCIR12, mTc UniCAMP01, mTcCIR40, mTc-UniCAMP04, mTc-UniCAMP03, mTc-UniCAMP02 (Lanaud, Risterucci, Pieretti, Falque, Bouet & Lagoda, 1999; Santos, Cerqueira-Silva, Mori, Ahnert, Correa & Souza, 2012) using the polymorphism information content (PIC) (Table 2). Primers were synthesized by Macrogen (Macrogen Humanizing Genomics) and were used for PCR amplification testing.
Amplification of microsatellite markers (SSR's)
For the PCR amplification, annealing temperature for each SSR~s was standardized using the available information in previous studies with each primer temperature for 40 s (Temperature decreased by 1°C for every cycle) (Table 2).
Different types of Taq polymerase were used to verify DNA performance as follows: GoTaq® DNA Polymerase (Promega) (Mix 1 (1 υ.μl-1), Taq polymerase (not commercial) (Mix 2 0.5 υ.μl-1), GoTaq® Green Master Mix (Promega) (Mix 3 (1 υ.μl-1)), and Taq DNA Polymerase recombinant (Thermo Scientific) (Mix 4 (1 υ.μl-1)) were evaluated. PCR reactions mixture (20 μl) containing 2.7 μL template DNA (10 ngμl-1), 1X PCR Thermo buffer, 2 mM dNTPs, 2.5 pmol each of forward and reverse primers, 2 mM MgCl2, 0.5 ng.ml-1 BSA, each of the Taq Polymerases described before and water to complete the total volume of the reaction.
Amplifications were carried out in a thermal cycler (LTCG Labocon 48-101). The amplified products were checked by agarose gel electrophoresis 3% after staining with ethidium bromide (0.5 mg.mL-1). Sizes were estimated by comparison with a DNA standard length marker (Gene ruler 50bp DNA HyperLadder® Bioline).
Results
Evaluation of developmental stages from leaves in cocoa field plant (FPL)
Three different protocols of DNA extraction were evaluated to determine their efficiency, from different leaves developmental stages. Stages C, D and E were selected, stage C are young leaves with a bronze coloration to light green, stage D is the transition between young leaves to mature leaves with a strong to light green and stage D are mature leaves with a dark green and leathery (Figure 1a).
A higher A260/A280 (>2) is indicative of RNA contamination, whereas lower A260/A280 (1.8) ratio is encountered when a contamination with protein occurs. When absorbance values ratio at 260/230 (<2), DNA could be contaminated by carbohydrates, salts or organic solvents (Demeke & Jenkins, 2010). The commercial DNeasy Plant Maxi Kit® failed to produce a good quality and high quantity of DNA, DNA concentration was no more than 25.80 ngμl-1 with lower quality A260/A280 (>2) and A260/A280 _(<1.8) ratios. CTAB Extraction method resulted in lower DNA yields with C leaves stage (5.27 ngμl-1 -2.38 A260/A280-0.28 A260/A230) and leaves of stage D (73.77ngμL-1; 60 A260/A280-1.03 A260/ A230). Leaves of stages D and E reaching higher concentrations, leaves of stage E was obtained with 136.77 ngμL-1 (1.77 A260/A280-1.15 A260/ A230) and with leaves of stage D plus sorbitol buffer was obtained 137.27 ngμL-1 (1.66 A260/ A280-1.66 A260/A230). DNA extractions with PowerPlant® Pro Kit resulted in highest DNA yields with all stages leaves. In leaves of stage D was obtained with 37.53 ngμL-1 (1.89 A260/ A280-1.22 A260/A230); stage D 71.77 ngμL-1 (1.91 A260/A280-1.66 A260/A230), following the C leaves stages 138.47 ngμL-1 (0.69 A260/A280 -0.20 A260/A230) and the stage D plus sorbitol buffer was 183.80 ngμL-1 (1.98 A260/A280-1.98 A260/A230) (Table 3).
Evaluation of DNA extraction in leaves of plant field and in vitro plantlets (IVL)
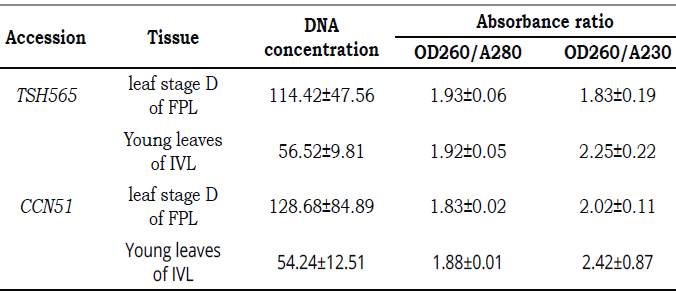
A protocol extraction for genomic DNA PowerPlant® Pro Kit is suitable for DNA leaf tissue of cocoa for both field and in vitro leaves. Highest DNA concentrations were obtained for FPL with 128.68 ngμL-1 and 114.42 ngμL-1 for CCN51 and TSH565 genotypes per 0.1 g of leaf tissue (Table 4). Subsequently, with IVL was obtained 54.24 ngμL-1 for CCN51 and 56.52 ng. μL-1 for TSH565, all results have allowed a high quantity with sufficient quality for later DNA applications (Figure 2).
Table 4 Quality and quality (OD260/A280-OD260/A230) of DNA isolated from field plants leaves (FPL) and in vitro plantlets leaves (IVL) of TSH565 and CCN51 with protocol for genomic DNA PowerPlant® with sorbitol buffer.
Data are means ±SE (n = 5).

Figure 2 DNA quality in agarose gel (0.8%) with protocol for genomic DNA PowerPlant® from FPL and IVL of TSH565 and CCN51. Numbered lanes from left to right with molecular marker (M: Gene ruler 1kb DNA HyperLadder® Bioline) (Cat no BIO-33053) IVL CCN51 (lanes 1-5), IVL TSH565 (lanes 6-10), FPL TSH565 (lanes 11-15), FPL CCN51 (lanes 16-20).
PCR amplification
Amplification conditions required the evaluation of different parameters in PCR reaction mixtures and these were modified according to positive amplification assays. The most important changes corresponded to different Taq polymerase enzyme that were used. With the mixture 1, 2, and 3 were obtained positive amplification using mTcCIR 17 SSR. However, the best result for this study was obtained with mixture 4, which is the one that uses the Thermo Taq polymerases enzyme (Figure 3).

Figure 3 PCR amplification of different Taq polymerase treatments for mTcCIR 17 microsatellite in agarose gel (3%) from FPL and IVL of TSH565. Numbered lanes from left to right with the molecular marker (M: Gene ruler 1kb DNA HyperLadder® Bioline), Mix 1 (GoTaq®) (Lanes 1 and 2), Mix 2 (Taq not commercial) (lanes 4 and 5), Mix 3 (GoTaq® Green Master Mix) (lanes 7 and 8), Mix 4 (Taq® Thermo Scientific) flanes 10 and 11) and negative control (NC) (Lanes 3, 6, 9, 12).
Despite of positive amplification obtained with mixture 4, an optimization was necessary to acquire a better define amplification band, by the fact that DNA concentration (1.75 ng.μl-1) had achieved a decreasing (1.35 ng.μl-1), in addition, have allowed an improvement in the volume of magnesium, which had decreased in a final concentration of 2 mM, to obtain a salt balance in final reaction concentration (Figure 4). In this sense, it was evident that quality and quantity of DNA obtained were sufficient for subsequent analyzes performed through SSR~s.

Figure 4 Amplification of different SSRs in agarose gel (3%) separation PCR products for FPL and IVL from TSHS565 and CCN51. Numbered lanes from left to right with the molecular marker (M: Gene ruler 50bp DNA HyperLadder® Bioline) (Lanes 1 and 37), mTc-UniCamp5 (lanes 1-4), mTc-CIR12 (6-9), mTc-UniCamp1 (11-14), mTc-CIR40 (16-19), mTc-UniCamp4 (21-24), mTc-UniCamp3 (26-29), mTc-UniCamp2 (31-34), negative controls (NC) (5, 10, 15, 20, 25, 30, 35).
Discussion
Secondary compounds in plants are the main problem encountered in DNA extraction and purification; these can damage the DNA and inhibit the action of the enzyme Taq polymerase. A major problem with DNA isolation from cocoa and most woody perennials is the contamination by polyphenols and polysaccharides, which interfere with DNA quality and downstream applications. To overcome this problem, we evaluated the influence of the leaf age of the plant material and different methods and additional wash with sorbitol to have achieve an improvement in contaminants removal.
Polyphenols are oxidized during nucleic acids extraction to form covalently quinones bond. Quinones irreversibly bind proteins and nucleic acids forming high molecular weight complexes, whereas polysaccharides co-precipitate with nucleic acids in low ionic strength buffers (Japelaghi, Haddad & Garoosi, 2011); thereby interfering with yield and quality of the isolated nucleic acids. These problems are further increased with plant age and/or unfavorable plant growth conditions.
Adult leaves (stage E) do not present high polyphenols content but have more mucilaginous channel. The presence of polysaccharides was quite evident in DNA extracted due to viscous aspect and glue-like texture; therefore, can coprecipitate DNA after alcohol addition, besides the contamination by polysaccharides makes DNA difficult to pipette and unamplifiable in PCR by inhibition of Taq polymerase activity, a result that was also founded.
In addition, stage D is the one that is characterized with a lemon shinning green that in this study is recognize like a transitional stage, which is characterized by having an intermediate concentration of chlorophylls, carotenoids, anthocyanins, polyphenols, etc. and therefore, a less quantity interfering substances. Also, during this transitional period the mucilaginous channels are not completely conformed because the full leaf expansion is yet sketchy carrying it out to have allowed a decreasing in the mucilage compounds. The extraction of stage D- leaf tissue from cacao genotypes results in an increasing yields of high-quality DNA with the PowerPlant® Pro kit protocol, including additional wash with a sorbitol buffer. The buffer was added to the kit with the objective to reduce the inhibitors and this is composed primarily of sorbitol, polyvinyl polypyrrolidone and β-mercaptoethanol (Souza et al., 2012). The sorbitol is used for removing mucilaginous polysaccharides, the reagent β-mercaptoethanol, which acts as a strong reducing agent, disrupts intramolecular disulphide bonds in proteins and polyphenols removal. Water-insoluble PVPP is used for phenolic compounds removal from plant extractions, since they form hydrogen bonds and subsequently, a precipitate is formed and removed from the plant isolation after centrifugation (Japelaghi, Haddad & Garoosi, 2011). Finally, DNA obtained was free of gelatinous substances, which usually coprecipitate with DNA, and it was free of proteins and phenols. The absence of phenolic compounds could be observed by the transparency of the DNA obtained. Cleaning the macerated tissue with the additional sorbitol buffer most of the polysaccharides and other contaminants that would otherwise hamper DNA extraction.
In the evaluated process of PCR conditions for the markers used was appropriate having previous information to reach adequate means of amplification, considering that a low concentration of enzyme ensures the fidelity of DNA amplification and a high concentration of enzyme, which results in amplification of unspecific products (Demeke & Jenkins 2010). Other factors that may modify the fidelity of DNA polymerase enzyme are the presence of 3'-5'exonuclease activity, the natural tendency of the enzyme to insert errors, the ease with which errors can be removed, salt concentration, specifically magnesium the one that showed to be a crucial factor in the activity of the Taq polymerase enzyme (data not shown). The reaction components such as DNA template, chelating agents present in the sample (EDTA or citrate), dNTPs and proteins can affect the amount of free magnesium. Its deficiency can inactivate the enzyme and reduce their excess and have allowed an increasing in non-specific binding (Schrader, Schielke, Ellerbroek & Johne, 2012). For these reasons, is essential to determine the optimal salt concentration, which generally is in the range of 1.5 to 3.0 mM.
The additive Bovine Serum Albumin (BSA) improves the quality of amplified products by PCR reaction, as concentrations above 0.8 μg.μl-1 of BSA, which results in an increasing of PCR efficiency, since acts as a getter ion and other protein inhibitors for Taq polymerase (Schrader et al., 2012). It also prevents the loss of enzyme such as adhesion of enzymes to reaction tubes and tip surfaces, BSA also improves specificity in amplification of regions with secondary structure, a reagent that was added also in this study to stabilize the final reaction and had achieved an increasing in PCR performance through DNA purity templates.
Amplifications of genomic DNA mold involve high quality strands multiplication as PCR methodologies. Which was confirmed by testing amplification through molecular markers type SSRs. It is noteworthy that no significant differences were found in the stage amplification, in accordance to the viability in DNA structure extracted in the types of plants used in vitro and field plant and their corresponding amplifications. Even though the performances observed in distinct Taq DNA Polymerase used, they seem to have slightly different sensitivities in PCR reaction, making the choice of enzyme in PCR a very important step to see the true prevalence structure of mold extracted.
Conclusion
This study presents a fast and efficient method for DNA isolation of high quality from cocoa leaves using a reduced amount of plant tissue. PowerPlant® Pro kit protocol was successfully optimized by additional wash with sorbitol buffer. The age of the plant material influences the efficiency and quality of the isolated DNA, being the stage D of developmental cocoa leaves in the field, which presented the best response. For the young leaves of the in vitro plants, the adjusted protocol was equally efficient in quality and quantity of DNA. On the other hand, is possible to conclude that the activity of Taq DNA Polymerase recombinant enzyme was highly effective compared with other enzymes of different commercial kits. Finally, The DNA obtained is being used for studies of somaclonal variation by SSRs markers.

















