INTRODUCTION
Microbial communities interact in different ways, either synergistically or antagonistically. To survive the adversities and coexist with other microorganisms, bacteria are continually fighting for nutrients and niche space (Hibbing et al., 2010). When it comes to bacterial antagonism, it is essential to define a producer strain as the one capable of producing toxic compounds that inhibit the growth of other non-producing strains, generally sensitive to the substance (Russel et al., 2017). Competition between bacteria can be influenced by the production of these toxic substances and producer strains are benefited compared to non-producing or sensitive strains by dominating the niche in which they are located (Khare and Tavazoie, 2015). However, producer and sensitive strains interact differently when they are in a structured environment than in an unstructured one (Kelsic et al., 2015; Chacón et al., 2018). In an unstructured environment where a population of sensitive strains has been established, producers are not able to invade because they pay the price for toxin production (i.e., energetic cost of plasmid carriage, production, and resistance to the molecule), decreasing their growth compared to the growth experienced by the sensitive strains. In a structured environment such as the surface of an agar plate, producers and sensitive strains grow in separate colonies and toxins diffuse from the producing colony towards the sensitive neighbors making resources more available to the producer strains, due to their excessive accumulation. Therefore, producer strains numbers increase compared to the sensitives, even if their growth rate is lower (Stubbendieck et al., 2016).
Different inhibitory substances produced by bacteria have been reported. Some of them include broad-spectrum antibiotics, organic acids, siderophores, and volatile organic compounds, antifungals, bacteriocins, among others (Riley, 2009; Li etal., 2013; Meena and Kanwar, 2015; Sindhu et al., 2016). Several inhibitory substances have not been elucidated yet; in fact, we only know 1 % of the bacterial diversity in a natural environment, leading us to assume that many other inhibitory substances remain to be explored. In this review, we will begin by describing the principal methodologies used to evaluate the production of these inhibitory substances. Next, we will provide some examples of diverse bacterial inhibitory substances including (i) bacteriocins, (ii) siderophores and (iii) other metabolites such as broad-spectrum antibiotics, covering their structural characteristics and modes of action. Finally, given the importance of the inhibitory substances for biotechnological purposes, their applications, as well as the use of beneficial bacteria as bio-inoculants, will be discussed.
METHODS TO EVALUATE MICROBIAL ANTAGONISM
Different methods can evaluate microbial inhibition, the most used comprise the double-layer agar and simultaneous inhibition assays (Molina-Romero et al., 2017a; b). Assays in liquid media have also been frequently reported and represent a variant of simultaneous active interaction (Kreth et al., 2008). All those methods have been used to evaluate the antagonism among bacterial or fungal strains, however, to determine the antagonism against nematodes and viruses other methodologies have been developed; one example is the microscopic observation to evaluate the paralysis of nematodes with inhibitory substances and diminution of disease symptoms produced by virus when the plant was inoculated with a beneficial bacteria (Wong et al., 2016; Su et al., 2017a; b) . This review only describes the most common methodologies used to evaluate microbial inhibition.
Double-layer agar
In the double-layer agar method bacteria never interact between them; however, bacteria explored as sensitive should be able to grow in the presence of metabolites previously produced by the antagonistic strain grown on the first agar-layer (Mukherjee and Ghosh, 2014). The double-layer agar method consists of growing a producer strain on the surface of an agar-medium during 24-48 h. After incubation time, producer colonies are removed with a sterile glass slide and the remain cells are killed by exposing the glass Petri dish to the vapor of chloroform during 1.5 h. Plates are left in a laminar flow cabinet until the residual chloroform is evaporated and the second layer of soft agar inoculated with the indicator strain is poured over the first layer of agar, where the producer strain had grown previously. Plates are incubated at the optimal temperature for each microorganism analyzed. Inhibition halos formed in the upper layer are considered indicative of antibacterial activity (see Fig. 1).

Figure 1 Double-layer agar assay. In this process the producer strain is grown in the middle of a glass plate with a specific culture media (a). After incubation of 48 h bacterial colonies are removed (b) and killed under chloroform vapors (c). Once the remaining chloroform is evaporated (d), a double layer of soft agar (inoculated with an indicator strain) is poured (e). Once more the plates are incubated to look for an inhibition halo (f).
Simultaneous inhibition
In the simultaneous inhibition assay, both bacterial species are co-interacting all the time during the assay. For this methodology, overnight cultures of strains explored as sensitive are placed over the surface of an agar plate by the spread-plating method (Sanders, 2012; Molina-Romero et al., 2017b), and a 20 µl-drop of the producer strain is placed on the middle of the agar plate. After the drop dried, Petri plates are inverted and incubated at the right temperature for the microorganism analyzed. Surrounding halos of the producer strain are indicative of antibacterial activity (see Fig. 2).

Figure 2 Simultaneous inhibition assay. An indicator strain is massively grown on the surface of an agar plate and a drop of the producer strain is placed in the middle of the plate. Once the drop dried, plates are incubated and the inhibition halo surrounding the producer strain is observed as shown in the last step.
Antagonism in liquid media
In this assay, the producer and the sensitive strains are grown in a defined liquid media, both separately and together in co-culture. Bacterial growth observed in the mixed culture is compared to the observed in the individual culture. When a producer bacterium inhibits the growth of a sensitive strain, the bacterial number of the sensitive strain decreases sharply in the mixed culture. In this experiment, the bacterial number is determined by counting CFU/ml using a selective medium, the media selection play an essential role in the screening of co-interacting strains (Muñoz-Rojas et al., 2005).
It is essential to highlight that the production of an inhibitory substance could be different in each bacterial growth conditions, growth phase and the kind of culture media used for this assay (Anacarso et al., 2014). This could be because gene expression depends on environmental and nutrient-availability conditions (McArthur and Bibb, 2008). Though, some strains can produce their inhibitory substances constitutively independently of the growth conditions (Muñoz-Rojas et al., 2005).
INHIBITORY SUBSTANCES PRODUCED BY BACTERIA
The most studied bacterial genera capable of producing inhibitory substances are Enterococcus, Lactococcus, Streptomyces, Bacillus, Pseudomonas, Klebsiella, Escherichia, and Burkholderia, and several articles have been published (Khabbaz et al., 2015; Sekhar and Thomas, 2015; Tontou et al., 2016; Huo et al., 2018). In this section, we will focus on some inhibitory substances produced by beneficial bacteria.
Siderophores
Iron, unlike other elementary nutritional sources such as nitrogen, phosphorus, potassium, among others, is not freely available in host organisms and is, therefore, an important limiting factor for the growth of microorganisms. Production of siderophores confers producing microorganisms a competitive advantage over other bacteria in the environment, excluding them from their ecological niche (Beneduzi et al., 2012).
Siderophores are small molecules (< 1500 Da) produced under iron-limited conditions and secreted to chelate iron from the environment. By diffusion, siderophores can attract the ferric ion with high affinity into the cell and also return it to the cell surface (Aguado-Santacruz et al., 2012). These molecules are mainly produced by Gram-negative bacteria, fungi, yeast and some graminaceous plants (phytosiderophores). Bacterial siderophores have been classified into different families according to their functional group: hydroxamates, catecholates, phenolates, and carboxylates. Additionally, there are some groups of siderophores that contain a mix of the main functional groups (Beneduzi et al., 2012; Ahmed and Holmström, 2014).
Some bacteria produce only one class of siderophores; however, other bacteria can secrete different types of siderophores, making them more efficient to colonize different environments. For example, some species of the genus Pseudomonas produce hydroxamates as ferribactine and pseudobactin, and other species produce molecules denominated pyoverdines of the type catechol (Pahari et al., 2017). About 270 siderophores have been structurally characterized, and their mechanism of transport has been described, and some variations have been found between Gram-positive and Gram-negative bacteria. For example, in Gram-positive bacteria, the Fe (III)-siderophore complex is bound to a periplasmic binding protein (that is anchored to the cell membrane) and eventually the complex is transported to the cytoplasm by ATP-dependent transporter systems. In contrast, Gram-negative bacteria carry out a more complicated process due to the presence of the outer membrane; involving TonB-dependent outer membrane receptors which recognize Fe (III)-siderophore complexes (Krewulak and Vogel, 2008). Once the complex binds to the outer membrane receptor at the cellular surface, it crosses the membrane through an energy-dependent mechanism carried out by membrane receptor proteins, periplasmic binding proteins, and inner membrane transport proteins; then, the complex is released into the periplasmic space and transported across the cytoplasmic membrane, where the complex is separated via reduction of Fe(III) to Fe (II) (Ahmed and Holmström, 2014).
It has been reported that siderophore-producing bacteria exert extensive biocontrol action against soil and root borne phytopathogens through the release of siderophores (Sah et al., 2017), Therefore, siderophore-producing bacteria protect plants from phytopathogens by acting as competitors, reducing the iron availability necessary for the pathogen growth (Beneduzi et al., 2012). Siderophore-producing bacteria also benefit plants by supplying them with iron when its availability is low in the environment, promoting plant-growth and improving phytoremediation (Chen et al., 2017). Siderophores produced by bacteria could be able to chelate other metals such as Cu, Cd, Ni, Zn, and others (Johnstone and Nolan, 2015; Chen et al., 2017). Moreover, bacterial siderophores have shown that protect plants by triggering induced systemic resistance (ISR) (Trapet et al., 2016). Some crops that have benefited from siderophore-producing bacteria include potato, sunflower, sorghum, oat, cotton, peanut, pigeon pea and cucumber (Dimkpa, 2016).
Several studies have reported the role of siderophores in biological control (Sayyed and Patel, 2011). Pseudomonas sp. strain B10 was the first bacteria showing biocontrol of plant pathogens. In particular, the synthesis of siderophores by fluorescent Pseudomonas promote plant growth and inhibit the growth of phytopathogens such as Erwinia carotovora, Ralstonia solanacearum, and Fusarium oxysporum. Pyoverdines, mainly produced by Pseudomonas aeruginosa and Pseudomonas fluorescens, have demonstrated to be adequate to control Pythium and Fusarium species. Pseudomonads also produce pyochelin, which is thought to contribute to the protection of tomato plants from Pythium, as reported in Pseudomonas aeruginosa 7NSK2 (Siddiqui, 2006).
It has been reported that some strains of Pseudomonas putida produce siderophores increasing yield and biosynthesis of the major essential oil components when they are inoculated to Mentha piperita (peppermint) (Santoro et al., 2015).
Burkholderia species are known to produce siderophores that inhibit multiple phytopathogens, for example, Paraburkholderia tropica (formerly Burkholderia tropica) (Tenorio-Salgado et al., 2013). Other strains such as Azospirullum brasilense have also shown biocontrol properties. This bacterium produces catechol type siderophores having in vitro activity against the fungus Colletotrichum acutatum, one of the most critical pathogens in strawberry crop (Tortora et al., 2011). Rhizobium species also produce catecholates, inhibiting the growth of fungal pathogens including Fusarium oxysporum, Fusarium solani, Ustulina zonata, and Fomes lamonensis. Indeed, using Rhizobium strains in pea production has helped to decrease the presence of Fusarium oxysporum in infested soils, improving crop growth (Siddiqui, 2006). Rhizobactin from Rhizobium meloti is another example; although it is not known if this siderophore participates in biocontrol, agronomically is very interesting since it allows the bacteria to be more competitive in the environment (Saha et al., 2016). Species of Azotobacter also produce different siderophores including aminochelin, azotochelin, protochelin and azotobactin which protect crops from pathogens such as Aspergillus, Alternaria, F. oxysporum, among others (Baars et al., 2015).
Bacteriocins
These molecules belong to the most abundant and diverse class of antimicrobial agents, constituting an unusual microbial weapon. At first, bacteriocins were defined as ribosomally synthesized peptides directed against bacteria closely related to the producer strain (Silva et al., 2018), differing from traditional antibiotics precisely to their "relatively" narrow spectrum; however, later in this review we will describe some examples of bacteriocins with broad-spectrum activity showing that beyond inhibiting only related strains, bacteriocins may inhibit other prokaryotes and also fungi or parasites (De la Fuente-Salcido et al., 2015). For this reason, we propose to define them as antimicrobial peptides that may or may not act on strains related to the producer bacterium, which may also be active against other bacterial genera, fungi and/or parasites.
It is believed that 99 % of all bacteria may produce at least one bacteriocin and the only argument why we do not know more bacteriocins is because they are poorly studied (Klaenhammer, 1988). Before studying a bacteriocin per se it is necessary to find a producer strain which is possible by performing antagonism assays (Muñoz-Rojas et al., 2005), and once the producer strain is found these molecules can be isolated and purified by diverse methodologies. We will notice that the chemical nature and biosynthesis mechanisms of bacteriocins may vary significantly so characterize a bacteriocin could become complicated. For example, in many studies it is only possible to obtain a partially-purified preparation because the activity or the concentration of the bacteriocin is lost after the purification steps (Gálvez et al., 2007); a bacteriocin recombinantly produced could be highly concentrated in only two steps of purification but the characterization may turn out unsatisfactory due to difficulties presented in the following assays, as was the case with the crystallization and X-ray assays of the bacteriocin LlpA (Parret et al., 2004). Consequently, in most of the cases, the structure or modes of action are not described, which encourages us to intensify the study of these intriguing molecules.
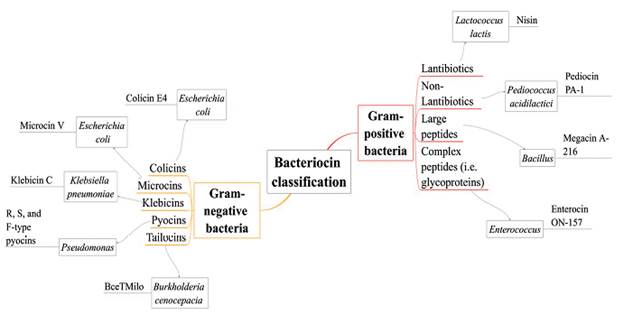
Several classifications of bacteriocins have been proposed; in this review we show them as bacteriocins from Gram-positive and Gram-negative bacteria (see Fig. 3), choosing the primary examples and mechanisms of action such as nisin, colicins, tailocins, pyocins, among others (Rebuffat, 2016; Silva et al., 2018).
Gram-positive bacteriocins
Bacteriocins produced by Gram-positive bacteria are generally cationic, amphiphilic, membrane permeabilizing proteins, with an approximate size ranging from < 5 to > 30 kDa. Several classifications of these bacteriocins have been proposed mainly according to their biochemical characteristics (Kemperman et al., 2003). The mechanism of action of Gram-positive bacteriocins is still under investigation, but it is accepted that they disrupt membranes through electrostatic interactions or by interacting with anionic membrane phospholipids causing pore formation (e.g. wedge-like and barrel-steve complexes), which results in the rapid efflux of the cytoplasmic compounds. In bacteriocins such as nisin and enterocin, it has been proposed that the antimicrobial activity is due to the presence of two structural domains (one located at N-terminus and one at C-terminus) where N-terminal rings play an essential role in binding the lipid II (main peptidoglycan transporter) preventing the correct synthesis of the cell wall (Gillor et al., 2009).
Members of the genus Bacillus are known to produce different bacteriocins, especially the lipopeptide type (Abriouel et al., 2011). The well-known bacteriocins produced by the genus Bacillus are subtilin and coagulin. Regarding this genus, Bacillus licheniformis ZJU12 was found to produce a bacteriocin-like peptide with a broad antagonistic spectrum. This peptide was able to inhibit the growth of some pathogenic microorganisms such as S. aureus, M. flavus, and some fungal phytopathogens such as Fusarium oxysporum; an interesting fact is that no adverse effects to mice have been detected in toxicity tests, which indicate a great prospect to use for biocontrol (He et al., 2006). Bacillus thuringiensis was considered as a model organism for producing antimicrobial compounds. Most of the bacteriocins synthesized by B. thuringiensis have a broad-spectrum, inhibiting phytopathogens such as Aspergillus and P. aeruginosa, being employed mainly for the control of plagues (Ugras et al., 2013; Salazar-Marroquín et al., 2016). In a recent study, it was reported a bacteriocin produced by an insect originated bacterium, Bacillus thuringiensis subsp. kurstaki Bn1; this bacteriocin was named as thuricin Bn1 and inhibits the growth of P. syringae, a plant pathogen (Ugras et al., 2013).
Gram-negative bacteriocins
Unlike bacteriocins from Gram-positive bacteria, Gram-negative bacteriocins are more extensive and carry out different mechanisms of action. One of the most known and extensively studied bacteriocin from Gram-negative bacteria is colicin, identified in E. coli(Riley and Wertz, 2002). Colicins are plasmid-encoded antimicrobial peptides secreted by E. coli and other related enterobacterial strains; their molecular weight varies between 20 kDa and 60 kDa and inhibit closely related strains such as Salmonella and other strains of E. coli. Production of colicins occurs mainly during times of stress like nutrient or oxygen depletion (Kaur and Kaur, 2015).
Structures of colicins are organized in three different domains: the translocation domain (T) N-terminally located, the receptor binding (R) located in the central region, and the cytotoxic domain (C) located at C-terminus, allowing to perform diverse mechanisms to kill bacterial cell (Cursino et al., 2002; Yang et al., 2014). Colicins target cells specifically through cell surface receptors so they can bind the outer membrane proteins by interacting with Tol or Ton complex periplasmic proteins and kill the sensitive strain, mainly through pore-formation, non-specific DNA degradation, murein and lipopolysaccharide biosynthesis inhibition (by interfering with lipid carrier regeneration), and inhibition of protein biosynthesis (Riley, 2009; Kaur and Kaur, 2015).
Other examples of bacteriocins produced by Gram-negative bacteria are microcins. These molecules are hydrophobic, low molecular weight, and ribosomally synthesized antimicrobial peptides. Their production is through a precursor peptide, including an N-terminal leader peptide and core peptides that may or may not undergo post-translational modifications. Microcins are characterized by showing heat, pH, and proteases tolerance, and do not require a lysis process to be secreted outside; indeed, they are secreted through the type I ABC (ATP binding cassette) transporter secretion system. Their mechanisms of action include pore-forming, DNase or RNase functions, and inhibitors of protein synthesis (Yang et al., 2014; Kaur and Kaur, 2015).
The genus Pseudomonas is also characterized for producing bacteriocins, well-known as pyocins. Pyocins target cells through specific receptors. Based on their structure, pyocins are classified as R, F or S-types. R-type pyocins are nuclease-protease resistant and it is thought that they have evolved from phage tails because their structure resembles non-flexible and contractile tails of bacteriophages. Their mechanism of action is through depolarization of the cytoplasmic membrane by pore formation. F-type pyocins are high molecular weight protease-resistant proteins which structure is similar to R-type pyocins, except for the flexible and non-contractile rod-like structure. S-type pyocins are colicin-like, protease-sensitive, and their structure consists in two components: the more significant component executes the killing activity (DNase, tRNase or channel-forming activities) while the smaller component, by showing sequence homology with colicin E2, is considered as an immunity protein. S-type pyocins cause cell death by DNA breakdown (pyocin AP41, S1, S2, S3) and pore formation (pyocin S5) (Michel-Briand and Baysse, 2002; Parret and De Mot, 2002).
Pyocins have shown a limited spectrum against other Pseudomonas species, in particular, they target Burkholderia cepacia complex strains; however, R-type pyocins have been shown to kill a diversity of P. aeruginosa strains as well as Campylobacter species, Neisseria gonorrhea, Neisseria meningitides, Haemophilus ducreyi, Pseudomonas fluorescens, and Pseudomonas putida (Naz et al., 2015). Another example is putadicin T01 produced by Pseudomonas putida which has shown a broad-spectrum against not only Gram-negative but also Gram-positive bacteria like Bacillus megaterium and Enterococcus faecalis, representing another opportunity for the treatment of pathogenic bacteria (Ghrairi et al., 2014).
Similar phage tail-like bacteriocins have been reported, particularly in plant-associated pseudomonad species. These molecules known as "tailocins", are large bactericidal structures with contractile (myotailocins) and flexible tails (siphotailocins) (Ghequire and De Mot, 2015; Yao et al., 2017). Both carry out a mechanism similar to phage infection: first, they reproduce the initial steps of the infection cycle by binding a cell receptor, and then the cytoplasmic membrane is punctured, where massive ion release occurs.
Recent studies have revealed that tailocins are not restricted to the genus Pseudomonas. For example, Burkholderia cenocepacia BC0425 produces a broad-spectrum tailocin (BceTMilo); it is suggested that BceTMilo binds to a D-glucose receptor for its adsorption through the Pseudomonas aeruginosa cell surface (Yao et al., 2017).
Other inhibitory compounds produced by bacteria
Bacteria also produce other metabolites such as volatile compounds and broad-spectrum antibiotics. Volatile compounds are vital in bacterial communication processes, but recent research indicates that these compounds, when secreted by bacteria, could perform antagonism over other microorganisms (Chaurasia et al., 2005; Kai et al., 2016). One of the volatile compounds produced by bacteria, mainly from the genus Pseudomonas, is hydrocyanic acid and it has been demonstrated that it participates in diverse antibiotic activities; evidence of this is the biocontrol of Thielaviopsis basicola (Matilla and Krell, 2017). Other volatile organic compounds (VOCs) produced by bacteria have been described; however, the biological role of most of them remains to be deciphered (Tyc et al., 2017).
Metabolites such as broad-spectrum antibiotics may have antibacterial and/or antifungal properties. Some of them include lipopeptides, generally produced by strains of the genus Bacillus. These molecules have been characterized by having an amphiphilic structure, which consists of the binding of a hydrophilic cyclic peptide to a fatty acid chain that can range from 12 to 14 carbon atoms (Meena and Kanwar, 2015). A very interesting feature of these molecules is the diversity of their structures which can influence their antimicrobial activity. In fact, their effectiveness to inhibit the growth of microorganisms has been proved, demonstrating that some of them only have activity against fungi, as is the case of iturines (Rojas-Solís et al., 2013); on the other hand, lipopeptides such as surfactins do have activity against bacteria and fungi (Meena and Kanwar, 2015). Other antibiotics described are the polyketides (PK) which contain in their structure multiple β-hydroxyketone or β -hydroxyaldehyde as functional groups. Some of them have a broad-spectrum antibacterial activity such as andrimid (Matilla et al., 2016) but others are only effective against fungi, for example, pyoluteorin and amphotericin B (Gomes et al., 2013; Matilla and Krell, 2017) (see table 1).
Table 1 Antibiotics produced by bacteria and their antimicrobial spectrum. Novel antibiotics with antibacterial, antifungal, antihelmintic and antioomycete activity are included.

A new family of antibacterial proteins was recently discovered, defined as lectin-like bacteriocins. These molecules are characterized by containing two carbohydrate-binding domains of the monocot mannose-binding lectin (MMBL) family. Some lectin-like bacteriocins from P. putida, P. syringae, and P. fluorescens were described: putidacin L1 or LlpABW from P. putida; LlpAPss642 from P. syringae; and LlpA1Pf-5 from P. fluorescens. These lectin-like bacteriocins can kill several Pseudomonas species but they are not active outside this genus (Parret et al., 2005). Similarly, the lectin-like bacteriocin LlpAXcm761from Xanthomonas citri pv. malvacearum LMG 761 can inhibit diverse species within the genus Xanthomonas (McCaughey et al., 2014).
PLANT-ASSOCIATED BENEFICIAL BACTERIA
In nature, interactions between microorganisms and plants may occur. For example, plant growth-promoting bacteria (PGPB) enhance plant growth through different mechanisms that include: direct mechanisms such as biological nitrogen fixation, phytohormone production, production of 1-Aminocyclopropane-1-carboxylate deaminase (ACC), phosphate solubilization and production ofvolatile organic compounds; and indirect mechanisms such as induction of systemic resistance (ISR), production of lytic enzymes and pathogen inhibition through the production of inhibitory substances (Lucy et al., 2004; Lugtenberg and Kamilova, 2009; Rojas-Solís et al., 2013; Molina-Romero et al., 2015). Other beneficial bacteria can degrade toxic compounds from contaminated soil, and several strains have shown the capability to biodegrade toxic compounds (Dvorak et al., 2017). It is noteworthy that inoculation of beneficial bacteria in plants could diminish the damage produced by chemical fertilizers on the environment and the cost of production (Baez-Rogelio et al., 2017; Pazos-Rojas et al., 2018). Moreover, beneficial microorganisms allow an increase in the size of roots, a better nutrient absorption, and diminution in the lixiviation level of combined nitrogen. Therefore, the addition of chemical fertilization could be diminished because the plant improves the efficiency to take the combined nitrogen (Dobbelaere et al., 2002a; Fuentes-Ramírez and Caballero-Mellado, 2005).
Depending on the colonization site of the plant, beneficial bacteria have been classified in rhizospheric, endophytic, epiphytic, bacteria from rhizoplane, and others. In the rhizosphere, region of the soil where diverse microbial communities live and are influenced by plant root exudates (Sylvia et al., 2005), bacteria are the most abundant microorganisms able to colonize and compete against the microflora of the roots, causing a neutral, detrimental or beneficial effect to the plant, specifically plant growth; these beneficial bacteria have been termed as plant growth promoting rhizobacteria (PGPR) (Vejan et al., 2016).
The main mechanism of the competition of PGPR is through the production of inhibitory substances (Beneduzi et al., 2012), resulting in an advantage for the elimination of phytopathogens.
ANTAGONISM OF RHIZOBACTERIA AGAINST PHYTOPATOGENS
Among the PGPR able to eliminate phytopathogens are included Gluconacetobacter diazotrophicus (Muñoz-Rojas et al., 2005), Azospirullum brasilense (Méndez et al., 2014), Pseudomonas fluorescens (Laue et al., 2000), Pseudomonas protegens (Ramette et al., 2011) and Burkholderia tropica (Bolívar-Anillo et al., 2016); and some Bacillus strains are also known for protecting plants from phytopathogens (Subramanian and Smith, 2015). For example, Gluconacetobacter diazotrophicus has shown in antagonism assays its ability to inhibit important phytopathogens such as F. oxysporum, F. solani, C. fimbriata and C. falcatum possibly due to the production of pyoluteorin (Logeshwaran et al., 2011).
Recently, it was described a new bacteriocin from Gluconacetobacter diazotrophicus, PAL5, named Gluconacin which has an antagonistic effect against phytopathogens such as X. albilineans (which produce leaf scald of sugarcane plants), and X. vasicola pv. vasculorum (causal agent of gumming disease of sugarcane and leaf streak of corn) (Oliveira et al., 2018).
Although Azospirillum is not well known as a typical biocontrol agent, some possible mechanisms to reduce damage by pathogens have been described; for example, the production of phenylacetic acid. One report also showed the ability of A. brasilense to produce siderophores with antifungal activity in vitro against Colletotrichum acutatum M11, preventing the anthracnose caused by this fungus (Tortora et al., 2011).
Some plant-associated Pseudomonas inhibit several phytopathogens such as Xanthomonas spp. (Garza-Ramos et al., 2015) and diverse bacteriocins produced by this genus have been isolated and characterized. For example, P. syringae pv. ciccaronei NCPPB2355 produces a bacteriocin that inhibits P. syringae subsp. savastanoi, the causal agent of olive knot disease (Lavermicocca, 1999); P. fluorescens strain BC8 produces the bacteriocin fluoricin-BC8 that inhibits P. solanacearum under in vitro conditions; Pseudomonas aeruginosa RsB29 cause suppression of Fusarium wilt and rot of chickpea (Sindhu et al., 2016); P. protegens CHA0 produces diverse secondary metabolites that include hydrogen cyanide, 2,4-diacetylphloroglucinol, pyoluteorin, and pyrrolnitrin to inhibit diverse phytopathogens such as Thievaloviopsis basicola and Pythium ultimum in tobacco and cucumber (Jousset et al., 2014; Sindhu et al., 2016).
Burkholderia species also inhibit essential phytopathogens. For example, Burkholderia tropica produces siderophores and volatile compounds that act as bio-controllers of phytopathogens such as fungi and nematodes, making it an excellent candidate to be used as a bio-inoculant in crops. The ability to inhibit the growth of phytopathogenic fungi by Burkholderia tropica was proven against Colletotrichum gloeosporioides, Fusarium culmorum, Fusarium oxysporum and Sclerotium rolffsi by producing 18 volatile compounds that included ce-pinene and limonene (Bolívar-Anillo et al., 2016). Burkholderia gladioli strains have also shown inhibitory activity in vitro and in planta against T. ptyseos, a pink disease causative agent (Marín-Cevada et al., 2012).
Bacillus strains such as Bacillus thuringiensis ssp. tochigiensis HD868 and Bacillus thuringiensis ssp. entomocidus HD9 (Subramanian and Smith, 2015) have been potentially accepted for protection against phytopathogens as Aspergillus niger, Aspergillus fumigatus, Aspergillus flavus, Cryphonectria parasitica, Fusarium oxysporum, Penicillium digitatum, among others. B. thuringiensis NEB17 produces the bacteriocin thuricin 17 (Th17) whose application in leaves soybean and corn stimulates the growth and it is the only bacteriocin studied extensively for plant growth promotion; it also participates as a bacterial signal compound and is able to increase phytohormones production and response to salt stress in Arabidopsis thaliana (Abriouel et al., 2011). B. amyloliquefaciens strain RC-2 produces a bacteriocin-like substance able to inhibit C. dematium and other phytopathogens such as R. necatrix, P. oryzae, A. tumefaciens, and X. campestris pv. campestris (Abriouel et al., 2011). Bacillus subtilis 14B reduced the percentage of infection in plants caused by Agrobacterium tumefaciens and it was proposed for biocontrol of crown gall disease in tomato plants (Hammami et al., 2009).
APPLICATION OF ANTAGONISTIC PGPB AS BIO-INOCULANTS
Beneficial bacteria have shown diverse potential functions when used in intensive agriculture, for example, plant growth. Furthermore, their ability to produce inhibitory compounds represents a potential for biological control. The strategy commonly used to kill phytopathogens is the use of chemical compounds such as pesticides. Nevertheless, reducing the use of pesticides is truly important since they have been implied in ecological, environmental and human health damages (Baez-Rogelio et al., 2017).
Although antagonistic interactions in rhizosphere for biocontrol purposes have not been intensively studied, it has been proposed that PGPR interactions with other soil microorganisms (fungal or bacteria) could be potentially used in plants of agricultural interest by developing mixed bio-inoculants or using their inhibitory substances per se (Ramamoorthy, 2001). Many reports have shown the biocontrol potential of PGPR, but most of the assays are only performed in vitro, and their direct application in crops has been little explored.
Mono-inoculants have been developed for commercial agriculture and are already being commercialized in several countries, mainly in Mexico and Argentina (Molina-Romero et al., 2015). One of the most explored bacteria in crops has been Azospirillum brasilense, which has been used in various crops showing successful results in more than 70 % of cases (Dobbelaere et al., 2002b). Other bacteria used for the development of these inoculants are Rhizobium etli, Pseudomonas fluorescens, Bradyrhizobium sp., Mesorhizobium cicerii, Sinorhizobium meliloti, Rhizobium legumonisrum biovar trifoli and Bradyrhizobium japonicum (Vivanco-Calixto et al., 2016).
The co-inoculation of microorganisms has already been reported and has apparently been more compelling, perhaps because of the synergistic effect that occurs when they are in co-interaction (Atieno et al., 2012; Zoppellari et al., 2014); for example, the co-inoculation of lettuce with Bacillus sp. and Glomus intraradices (Vivas et al., 2003) and co-inoculation of pea with Rhizobium and Bacillus megaterium (Elkoca et al., 2010). Few formulations containing more than three species of microorganisms in consortium have also been studied (Molina-Romero et al., 2015), one of them is the inoculation of sugarcane with a mixture of five diazotrophic bacteria ( Gluconacetobacter diazotrophicus, Herbaspirillum seropedicae, H. rubrisubalbicans, Azospirillum amazonense, Burkholderia tropica) (Oliveira et al., 2009). Although mono and co-inoculations have resulted excellent for crops, all these formulations have been marketed mainly to promote plant growth and not for biocontrol purposes; examples of this are Bradyrhizobium spp. or A. brasilense inoculants which have been commercialized for years to increase the yield of diverse crops (Fukami et al., 2016). For this reason, designing new formulations of mono or multi-inoculants with beneficial bacteria capable of eliminating phytopathogens is a challenge.
The design, formulation and optimization of a compelling mixture of bacteria to be used as inoculants is not an easy task; it requires studies of adhesion to seeds and colonization in plants (Sundaramoorthy et al., 2012; Singh et al., 2014; Baez-Rogelio et al., 2017). Also, antagonistic assays among the microbial strains of the mixture should be performed before the design and application of a multi-species inoculant, because some antagonistic effects could occur among bacteria (Molina-Romero et al., 2017b). It also requires assays to guaranty the coexisting of bacterial strains when they are in the formulation and associated with plants and verify the plant growth promotion effectiveness (Muñoz-Rojas et al., 2013). Several polymicrobial formations contain microbial strains capable of coexisting without antagonizing each other with the capability of eliminate pathogens (Oliveira et al., 2009; Muñoz-Rojas et al., 2013; Baez-Rogelio et al., 2017; Molina-Romero et al., 2017b; Pérez-Santos et al., 2017a; b) and some of these formulations contain desiccation-tolerant bacteria, making them more efficient in environments with low water availability (Molina-Romero et al., 2017b; Pérez-Santos et al., 2017a; Pazos-Rojas et al., 2018).
Some bacterial inoculants are commercially available for promoting growth in crops and are also useful for biocontrol; however, some of them inhibit pathogens due to the presence of fungicides (Crovo and Clemente, 2015) such as metalaxyl, fludioxonil or benomyl which may cause health damages.
Recently, in the Laboratory of Ecology and Survival of Microorganisms from the Center of Research in Microbiological Sciences (Sciences Institute, University of Puebla, Puebla, Mexico) multispecies inoculants have been developed, fulfilling the challenge of containing strains capable of coexisting without antagonizing each other and also able to eliminate phytopathogens (Muñoz-Rojas et al., 2013). Other examples of mono and multi-inoculants that inhibit the growth of phytopathogens include Enerbac from the company "Agrícola Inovación-Mexico", Fungikillerâ from Bio-Iliberis R&D, and Serenade ASOâ from Bayer CropScience, among others (Matilla and Krell, 2018).
Microbes often exist in complex multispecies communities in the environment, but the molecular mechanisms through which such communities develop and persist, despite significant antagonistic interactions between species, are not well understood (Wong et al., 2016). It would be interesting to perform research related to the effect of beneficial antagonistic bacteria on rhizosphere bacterial communities and evaluate ifmultispecies formulations influence is stronger than mono-inoculants. Plant microbiomes are fundamental to understanding how to improve the health of plants and crops production (Berg et al., 2014; Busby et al., 2017). In this context, studies of microbial diversity are critical to the prevention of diseases and can be implemented as a biomarker in plant protection strategies (Berg et al., 2017). An effective biocontrol should be based on the knowledge of the microbiomes present in healthy and thriving plants.
CONCLUSIONS
By producing diverse antimicrobial compounds, beneficial bacteria play a very important role in different areas, particularly in agriculture. The use of these microorganisms as bio-inoculants represents a great strategy to fight against phytopathogens since their production is cheaper than any other chemical fertilizer and they have positive effects on plants. Moreover, reducing the use of chemical fertilizers and toxic compounds such as pesticides and herbicides is a critical factor to preserve the environment and human health. Although many bacterial inoculants have been designed, marketing formulations with bacterial consortiums, especially those with the capability to inhibit the growth of diverse kind of pathogens, is still in development. Having them available will contribute to sustainable agriculture by reducing the use of toxic compounds without affecting agricultural productivity.