Introduction
Glyphosate is the most used herbicide worldwide. It is a broad-spectrum herbicide that acts by inhibiting the enzyme 5-enolpyruvyl-shikimate-3-phosphate synthase or EPSPS (EC 2.5.1.19) (Duke, 2018). This enzyme is a key element of the shikimate pathway, by which plants produce metabolites derived from chorismate, including the aromatic amino acids tyrosine, phenylalanine, tryptophan, and physiologically important compounds such as pABA and Coenzyme Q10 (Tzin and Galili, 2010). Specifically, EPSPS catalyzes the synthesis of 5-enolpyruvyl-shikimate-3-phosphate (EPSP) from shikimate-3-phosphate (S3P) and phosphoenolpyruvate (PEP) (Maeda and Dudareva, 2012). Given the structural similarity between glyphosate and PEP, the first acts as a competitive inhibitor of EPSPS, impeding the reaction it catalyzes (Funke et al., 2006), and therefore, hindering plant growth and eventually causing its death.
EPSPS is present in plants, bacteria and fungi, but not in animals, which do not synthesize their own aromatic amino acids (Padgette et al., 1995). There are several natural variants of EPSPS that are not affected by glyphosate; most are of bacterial origin (Barry et al.,1997; Yan et al., 2011; Cao et al., 2012), and some have been reported in plants (Mao et al., 2016). Some of these variants have been used to generate glyphosate-tolerant transgenic plants, and the EPSPS from the Agrobacterium tumefaciens strain CP4 (Barry et al., 1997) is the most commonly used in commercial transgenic crops.
CP4:EPSPS is a class II EPSPS enzyme with high enzymatic efficiency and tolerance to inhibition by glyphosate. It also shows a low Km (Michaelis constant) for PEP [12 μM] (Barry et al., 1997), indicative of a high affinity for this substrate. Barry et al. (1997) identified several class II EPSPSs, of which CP4:EPSPS showed the best kinetic parameters. Its tolerance to glyphosate is partly explained by the presence of an A100G residue in the enzyme's active site. Funke et al. (2006) restored CP4:EPSPS glyphosate susceptibility by means of an Ala100-Gly100 substitution which generates a slight change in its three-dimensional structure that allows the binding of glyphosate in an extended conformation that inhibits the enzyme. The presence of an A100G residue generates a narrower active site that only allows the binding of glyphosate in a condensed conformation that does not inhibit the enzyme (Funke et al., 2006).
A transgenic plant with a functional cp4:epsps gene in its genome is glyphosate-tolerant because the presence of CP4:EPSPS is enough to ensure the synthesis of the required aromatic amino acids. In contrast, conventional plants when exposed to commercial doses of the herbicide show a marked reduction in growth and eventually they die (Padgette et al., 1995).
One of the most important issues related to the production of a glyphosate-tolerant transgenic plant is to ensure good expression levels of the transgene. Gene sequence modifications as codon usage or guanine-cytosine (GC) content may improve transcription levels and protein production (Chou and Moyle, 2014; Jeong et al., 2017; Sivamani et al., 2019).
In addition to biologically relevant aspects for the design of the expression cassette, our work also considers that the genetic elements used have freedom to operate in Colombia. A Freedom to Operate (FTO) analysis is an evaluation of the intellectual property (IP) aspects involved in a research project or in the development of a product to make sure it can be carried out with a low or tolerable probability of infringing current patents or other IP rights (Bennett et al., 2008). Performing this analysis is vital during the first stages of the research process if it involves commercial interests (Mora-Oberlaender et al., 2018) such as in the present study.
Here we present the in silico design of three expression cassettes for conferring glyphosate tolerance to plants. The sequence of the cp4:epsps gene was optimized towards soybean codon usage. In our approach, the cp4:epsps gene is both the gene of interest and the selection marker. The objective of this study was to assess the functionality of the expression cassettes by transforming the model plant Nicotiana benthamiana. Additionally, in order to advance the development of herbicide tolerance agbiogenerics, we carried out an FTO analysis for the elements of the expression cassettes in order to establish if their eventual commercial use would affect third party rights.
Materials and methods
In silico design of expression cassettes
Three different expression cassettes that would confer glyphosate tolerance were designed and denominated as E-IGP, E-IGP2, and E2. The coding sequence used in the three cassettes corresponds to that of the cp4:epsps gene. As promoters we used either the soybean polyubiquitin, Gmubi, (Chiera et al., 2007), the single (Odell et al., 1985) or the 1 duplicated (Kay et al., 1987) version of the CaMV35 promoter (Fig. 1).
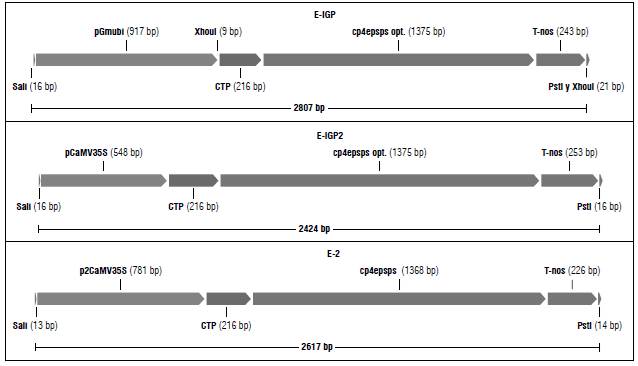
FIGURE 1 Schematic diagram of the three expression cassettes designed using Gene Designer 2.0 (Villalobos et al., 2006).
In plants, the epsps gene is found in the nuclear genome. When expressed, EPSPS is transported to the chloroplast by means of a transit peptide, CTP, which is then removed by a site-specific metalloprotease (Della-Cioppa et al., 1986). Therefore, we included a signal sequence, CTP, in the 5' end of the coding sequence.
The sequences for the coding regions of the expression cassettes were obtained from published patents. Specifically, the cp4:epsps gene and CTP transit peptide from Petunia hybrida correspond to SEQ ID 9 (gi:2469099) and SEQ ID 14 (gi:2469102), respectively, from patent US 5633435 (Barry et al., 1997). The sequences of the promoter regions were downloaded from NCBI, where the Gmubi promoter corresponds to gi:162280984 and the CaMV35S promoter corresponds to gb|HQ698853.1|:1967-2514.
The designed expression cassettes were based on a previous study (Jiménez and Chaparro-Giraldo, 2016). The coding sequence or open reading frame (ORF) of the cp4:epsps gene was optimized according to the following criteria using the software Visual Gene Developer-VGD 1.3 (Jung and McDonald, 2011): modifications for soybean codon usage, removal of cryptic splice sites and of premature polyA sites. The final optimized sequence was selected according to parameters such as the codon adaptation index (CAI) (Sharp and Li, 1987), effective number of codons (Nc) (Wright, 1990), and GC content. All modifications of the nucleotide sequence were synonymous, leaving the original amino acid sequence of the coded protein unchanged. Gene and regulatory sequences were assembled into expression cassettes, and restriction sites were introduced using the software Gene Designer 2.0 (Villalobos et al., 2006). We carried out an in silico translation analysis on a primary structure level. NCBI's ORF Finder and Blast-x were used to predict the expressed amino acid sequence.
Each of the three expression cassettes designed and evaluated in silico were synthesized and cloned into a pCAMBIA 1301 vector from which the plant-selection gene and reporter gene were removed. They were then used to transform cells of A. tumefaciens, strain LBA4404, by electroporation. Transformed bacterial cells were selected phenotypically and evaluated by PCR.
Genetic transformation of Nicotiana benthamiana
In order to use glyphosate as a selection agent for transformed plants, we measured the sensitivity of Nicotiana benthamiana to different concentrations of the herbicide. Leaf explants were placed on R media (1X MS salts, 1X Gamborg vitamins, 30 g L-1 sucrose 1 mg L-1 BAP, 7 g L-1 agar PTC, and pH 5.8) with increasing concentrations of glyphosate: 0, 10, 25, 50, 100, 250, 500, 1000, and 2000 uM. These concentrations were chosen accordingly to what has been reported for the closely related species Nicotiana tabacum (Wang et al., 2003; Fathi-Roudsari et al., 2009; Akbarzadeh et al., 2010; Yan et al., 2011). The herbicide (tissue culture grade N-(Phosphonomethyl) glycine, Phyto-Technology®) was added to the growth medium prior to pH adjustment.
Transformation of N. benthamiana was mediated by A. tumefaciens. Bacteria were grown on liquid LB medium with acetosyringone (200 μM), kept at 28°C and shaken at 200 rpm until they reached an optical density (OD) of 0.6. Square-shaped (approximately 1 cm2) leaf explants were used for infection by placing them in liquid coculture medium for two minutes with a bacterial suspension and then transferred to solid coculture medium for 24 h (R medium with 200 μM acetosyringone). Explants were then washed five consecutive times at room temperature and 100 rpm in order to remove bacteria. They were then dried on sterile paper towels and placed on selective medium 1 (R medium with 500 mg L-1 carbenicillin). After two weeks, explants were transferred to selection medium 2 (R medium with 500 mg L-1 carbenicillin and 10 μM glyphosate). Regenerants were placed in propagation medium (1X MS salts, 1X Gamborg vitamins, 30 g L-1 sucrose, 2.5 g L-1 Gelzan, and pH 5.8) and kept in vitro under controlled photoperiod (16/8) and temperature (28°C) until they were used for molecular tests. Untreated explants were used as controls. Absolute control explants were placed on R medium, and negative control explants were placed on selective medium. All assays were carried out with four-week-old plants.
Molecular tests
Potential transformants of N. benthamiana were evaluated by conventional PCR. We designed specific primers for the version of the cp4:epsps gene in each expression cassette. Plants potentially transformed with cassettes E-IGP or E-IGP2 were tested with primers E-IGP1FW: TCAC-CATGGGGCTTGTAG and E-IGP1RV: GCTATACGGT-GATCGAGATGC. PCR conditions used were an initial denaturation cycle (95°C x 10 min) followed by 35 amplification cycles (95°C x 60 sec, 61°C x 90 sec, 72°C x 90 sec) and a final elongation (72°C x 5 min). For the E-2 cassette we used primers E-2FW: ATATCCGATTCTCGCTGTCG and E-2RV: CCATCAGGTCCATGAACTCC. Here, we used Kapa Biosystems polymerase and the PCR conditions were one initial denaturation cycle (95°C x 6 min) followed by 55 amplification cycles (95°C x 20 sec, 66 °C x 15 sec, 72°C x 40 sec) and a final elongation (72°C x 30 sec). Amplification products were visualized by agarose gel electrophoresis stained with Ethidium bromide
Plants that showed a PCR product of the expected band size were selected for RT-PCR analysis. Total leaf RNA was extracted using Norgen Bioteck Corp. RNA/DNA/ protein purification kit (Thorold, Canada). DNA contamination was eliminated with the DNaseI RNA free kit from ThermoScientific (Waltham, Massachusetts, USA), and its effectiveness in removing DNA was tested by conventional PCR for the constitutive actin gene using primers FW:TGGTACAAGGGTCCATAGCG and RV: GCCGTCCTCTCTCTGTATGC. These primers generate a 518 bp amplicon. cDNA synthesis was performed using the First strand cDNA synthesis kit from ThermoScientific (Waltham, Massachusetts, USA), and its quality was checked using the actin primers in a PCR assay. Expression of the genes in E-IGP, E-IGP2 and E-2 was evaluated by PCR assays using the primers described above.
For all PCR assays, plasmid DNA was used as positive control, DNA from a non-transformed plant was used as negative control, and the reaction mixture without DNA was used as an absolute control.
PCR positive individual plants (4-5-week-old) were hardened for phenotypic evaluation. They were transferred to a 3:1 peat: soil mixture and kept under controlled growth conditions as above. Plants kept under these conditions for one month were used for evaluation by applying 0.2% glyphosate on the entire aerial plant surface. The assay was performed in duplicate. The outcome of the test was evaluated 15 and 30 d after herbicide application.
Freedom to Operate Analysis (FTO)
In order to establish the potential for the eventual commercial use of transgenic plants transformed with the expression cassettes described here, we performed an FTO analysis of the genetic elements they included. This analysis was limited to Colombia. First, a patent search was carried out in three international patent databases, The Lens (https://www.lens.org/lens), Patentscope (http://www.wipo.int/patentscope/en/), and Spacenet (http://www.epo.org/). The search was then performed in the database of the Colombian Superintendence of Industry and Commerce (SIC), which is the national authority for patents (http://www.sic.gov.co/es/banco-patentes). All these databases are publicly available at no cost. In the international patent databases, we used key terms to search within the claims to identify patents related to the genes and regulatory sequences used. Once identified, the national database was queried using key terms to search within the relevant fields (inventor, assignee and title). All retrieved documents were analyzed to determine if the gene sequences or regulatory elements are protected by IP rights in Colombia. The analysis was updated to November 2019.
Results and discussion
In silico design of expression cassettes
Previous research has shown the use of glyphosate as a selection agent in the genetic transformation of plants from several species like rice, maize, cotton and soybean, using concentrations in a range from 0.5 mM to 10 mM of the growth medium (Latif et al., 2015; Ren et al., 2015; Soto et al., 2017). Most reports of genetic transformation of N. tabacum to confer tolerance to glyphosate use antibiotics to select transformed plants (Wang et al., 2003; Yan et al., 2011; Peng et al., 2012), but there is evidence of the use of glyphosate to select transformants in this species (Akbar-zadeh et al., 2010). Here, we designed synthetic versions of the cp4:epsps gene in order to use it as both the gene of interest and a selectable marker. The different versions of the gene were included in three expression cassettes, E-IGP, E-IGP2 and E-2. The first two included the gene with modifications for codon-usage optimization in soybean in order to enhance its expression in that species. The third expression cassette carried the native sequence of the gene.
Codon-usage modification has been shown to be one of the most important factors for obtaining good levels of expression for a heterologous gene (Yan et al., 2011; Kucho et al., 2013; Sivamani et al., 2019). Codon bias is usually measured by CAI (Sharp and Li, 1987). Most genes used in genetic transformation in published research are modified to favor the codon usage of the transformed species, but a few studies give the CAI or the value of some other parameter for the modified sequence. Kucho et al. (2013) improved the translation efficiency of the gene for gentamicin tolerance using a modified version with a CAI of 0.835. A similar effect was reported for the expression of the Toxoplasma gondii SAG1 antigen in tobacco, in which a CAI of 0.83 was obtained (Laguia-Becher et al., 2010). Yan et al. (2011) found that modified sequences of the Pparo1 gene with CAI values of 0.7 and 0.9 conferred good tolerance to glyphosate in transgenic tobacco plants. Accordingly, we modified the coding sequence of the cp4:epsps gene until a CAI of approximately 0.8 was achieved. In total, 325 codons (71.42%) from 455 codons of cp4:epsps coding sequence were optimized, where the synthetic gene sequence was 74% identical to the native gene sequence.
Genetic expression is a complex process and codon usage is not the only factor determining its efficiency (Jung and McDonald, 2011). During gene design it is also important to consider the GC percentage of a sequence, a factor that is related to codon usage. In plants, GC content is higher in monocots than in dicots, especially when considering the third base of each codon [GC3] (Clément et al., 2014; Singh et al., 2016). By optimizing the coding sequence, Li et al. (2013) were able to improve expression levels of the gene cry1Ah and resistance to insects in plants. Through codon-usage modifications they changed the GC content from 37-48% (native gene) to 55-63% (designed genes) and found greater expression of the designed genes both at the mRNA and protein levels, which led to a better resistance to insects. The unmodified cp4:epsps sequence has a high GC content (66%), especially in GC3 (84.21%). This value is notably higher than that reported for plant genomes (Singh et al., 2016). Using Visual Gene Developer 1.3, we changed the total GC content of the gene sequence to a final value of 51.78% and we changed the GC3 content to 48.21%. The percentage of GC3 is the most notable factor in discriminating codon usage between monocots and dicots. In the first group, 16 out of 18 amino acids favor G or C in the third position, while in dicots this number is 7 out of 18 amino acids (Murray et al., 1989). A high GC3 content in mRNA codons also leads to a higher potential for the formation of hairpin structures, which can affect the expression and stability of mRNA (Barry et al., 1997). Given this, reaching a GC3 content of 48.21% of cp4:epsps seems appropriate for its expression in soybean and other dicots. The total GC content in cassette E-2 (unmodified cp4:epsps gene) is 54.58%, while the cassettes E-IGP and E-IGP2 reached 44.64% and 47.36%, respectively, closer to the value reported for soybean (46%).
The effective number of codons (Nc) (Wright, 1990) measures the degree of synonymous codon usage bias with a number ranging between 20 and 61. If Nc is 20, codon usage is extremely biased, while an Nc of 61 corresponds to a homogeneous use of synonymous codons. The original cp4:epsps sequence has an Nc of 27, which is a relatively biased codon usage. In the modified version, we obtained an Nc of 49.5.
Every change made in a DNA sequence, even if it is to make it closer to the codon usage pattern of the host plant, can lead to new unexpected signals, such as cryptic splice sites or premature polyA sites. Any such sites were eliminated by synonymous codon changes that did not alter the reading frame. Figure 1 shows a diagram of the expression cassettes designed as described above. By in silico translation, using ORF finder and Blast-x, we checked that these cassettes effectively code for the CP4:EPSPS protein.
Genetic transformation of Nicotiana benthamiana
We evaluated the effect of glyphosate on the in vitro regeneration of N. benthamiana. In a non-treated control, the first sprouts were visible four weeks after planting and they were present in 98% of explants (Fig. 2). Explants planted under the lowest evaluated glyphosate concentration (10 μM) had a similar regeneration percentage, while at 25 there was a drastic reduction (11.4%). In a concentration of glyphosate equal or above 50 we observed no regeneration. In the selection of transgenic N. tabacum plants, Akbarzadeh et al. (2010) use 0.1 mM glyphosate. Our results show that for N. benthamiana this concentration not only inhibited regeneration but was lethal for the explants.
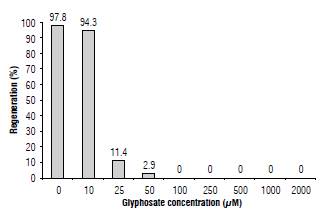
FIGURE 2 Regenerative response of N. benthamiana explants under different glyphosate concentrations.
The three expression cassettes we designed were used for independent genetic transformation assays of N. benthami-ana. Initially we used 25 μM glyphosate for the selection of possible transformants, but most régénérants had an abnormal phenotype (data not shown). Fathi-Roudsari et al. (2009) compared two different selection strategies using glyphosate in N. tabacum. The authors found better regeneration results when using a glyphosate-free medium for two weeks and then an initial selection concentration of 5 mM, which they doubled every two weeks up to a lethal dose of 50 mM. Accordingly, we used the two-week incubation period with no herbicide present and then transferred the expiants to a medium with 10 μM glyphosate. We observed the first régénérants between weeks three and four.
Transformation assays with the expression cassettes E-IGP, E-IGP2 and E2 were set up with 80 expiants each. We obtained 18 (22.5%), 37 (46.25%) and 25 (31.25%) régénérants, respectively. These values are notably lower than the regeneration percentage (95% average) for control expiants not subjected to transformation or selection with the herbicide, showing the negative effects of the transformation and selection process on regeneration. The two-week incubation period without the selective agent was effective in reducing the amount of abnormal régénérants. Most of the co-cultured expiants produced normal régénérants, while those used as negative control (non-transformed expiants placed in identical selection conditions) generated mostly abnormal régénérants.
Molecular tests
Plants that were phenotypically selected as possible transformants were evaluated by PCR. The expected size of the amplified fragment generated by the primers designed to detect the cp4:epsps gene in the expression cassettes E-IGP and E-IGP2 is 907 bp. This fragment size was effectively detected in three plants transformed with E-IGP and six with E-IGP2 (Fig. 3). For those plants transformed with E-2, the expected fragment of 341 bp was detected in seven individuals. Despite losing some of the initial explants due to A. tumefaciens contamination, the transformation efficiency for each of the expression cassettes was 16.6% (E-IGP), 16.2% (E-IGP2) and 28% (E2). Glyphosate has been used successfully as a selective agent for transformed plants in cotton (Latif et al, 2015), soybean (Soto et al, 2017), maize (Ren et al, 2015), and other species. We obtained transformed N. benthamiana plants using this selection approach, but it was with a low transformation efficiency, possibly due to the effect of the herbicide on the regeneration capacity of the explants.
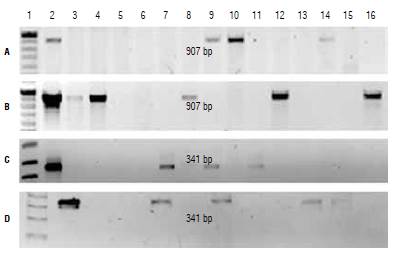
FIGURE 3 PCR identification of transformed plants. PCR product from plants transformed with A) E-IGP cassette, B) E-IGP2 cassette, and C and D) E-2 cassette. 1, molecular weight marker; 2, positive control; 3-16, PCR product from evaluated plants.
In most plants for which a positive PCR result was obtained, we also detected the presence of a primary transcript by RT-PCR (Fig. 4). This suggests that the expression cassettes in these plants are functional. In two plants in which we detected the presence of the transgene by PCR (each one from cassettes E-IGP2 and E-2), there was no evidence of cp4:epsps mRNA by RT-PCR. Different factors may account for this result such as a position effect, since the transgene may have integrated into a heterochromatic region of the genome or in highly repetitive regions that may hinder expression (Kohli et al, 2006). Other factors include the possible silencing of the transgene due to a multiple number of copies integrated into the genome (Velten et al., 2012; Khuong et al, 2013) or methylation processes (Rajeevkumar et al, 2015). Regarding the last issue, the 35S sequence used in two of the expression cassettes has been associated with silencing by methylation (Okumura et ah, 2016; Shimada et ah, 2017; Wang et ah, 2017). Additional research would be needed in order to determine the cause of the absence of cp4:epsps mRNA in these two plants.
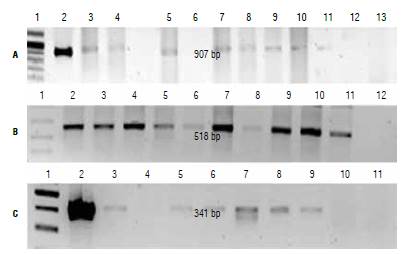
FIGURE 4 RT-PCR assay of transformed plants. A) Plants transformed with cassettes E-IGP and E-IGP2, primers: E-IGP 1, molecular weight marker (MW); 2, positive control; 3-5, plants transformed with cassette E-IGP; 6-11, plants transformed with cassette E-IGP2; 12, negative control; 13, absolute control. B) Amplification of endogenous gene with Actine primers. 1, MW; 2-11, evaluated plants; 12, absolute control. C) Plants transformed with cassette E-2, primers E-2.1, MW; 2, positive control; 3-9, evaluated plants; 10, negative control; 11, absolute control.
Phenotypical evaluation
PCR-positive plants that were successfully hardened and established were evaluated phenotypically to further test the functionality of the expression cassettes. Glyphosate (0.2%) was applied on all the shoots. In non-transformed controls, wilting was evident 8 d after treatment and death occurred 15 d after treatment. Transformed plants showed no negative effects during this period (Fig. 5). These results, together with the molecular evidence of the presence and transcription of the transgene, indicate that the cassettes we designed effectively confer glyphosate tolerance to transformed plants.
Freedom to Operate Analysis
An FTO analysis is a valuable tool which could be used as a strategy towards the development of agbiogenerics. An FTO analysis of a genetically modified crop should include all the elements involved in its obtention, such as expression cassettes, vectors, plant material, laboratory protocols, etc. (Mora-Oberlaender et ah, 2018). As a first step in this process, an FTO analysis of the designed expression cassettes was performed. The patents related to the sequences of the different genetic elements were identified and analyzed. The most relevant ones are summarized in Table 1. Our search yielded several patents in the United States, most of which have already expired. The sequence of the gene cp4:epsps (patent US 5633435) became part of the public domain in 2014 and its patent was never requested in Colombia. No requested or assigned patents for the promoter regions we used were found in the Colombian jurisdiction. The only patent directly related to the subject of interest is in the SIC database under file number 07136194. This patent protects the use of a sequence that includes all or part of the expression cassette in soybean event MON89788 and its flanking sequences, which, in other words, protects the event itself. Therefore, this patent does not affect the use of the sequences used in the expression cassettes we designed and described here.
Conclusions
We designed three expression cassettes (E-IGP, E-IGP2 and E2) that included the gene cp4:epsps and conferred tolerance to glyphosate to transformed plants. Two of these cassettes (E-IGP and E-IGP2) included a codon-usage modification to favor expression in soybean. The functionality of these cassettes was evaluated by genetic transformation of the model plant N. benthamiana and the phenotypical testing of transformed lines as well as molecular assays such as PCR (to determine presence or absence of the transgene) and RT-PCR (to detect transcription). We detected the presence and transcription of the transgene as well as tolerance to the herbicide in plants transformed with each of the three expression cassettes. This suggests that they are all functional and could be used further in genetic transformation of plants. The FTO analysis we performed suggested that the potential commercial use of these cassettes does not infringe third-party rights in Colombia. This analysis, however, must be validated periodically before commercial use.
















