Introduction
Camu-camu (Myrciaria dubia (Kunth) McVaugh) is a perennial species native to the Amazon basin that stands out for having high concentrations of vitamin C (7.36 g/100 g of pulp) (Chagas et al., 2015) and other antioxidant compounds, such as ellagic acids, ellagitannins, proantho-cyanidins, epicatechin, catechin, delphinidin 3-glucoside, cyanidin 3-glucoside, and rutin (Chirinos et al., 2010; Fracassetti et al., 2013; Fidelis et al., 2020). The pulp can be processed into carbonated drinks, concentrated juices, vinegar, ice cream, candies, pills, energy drinks, creams, colognes, among others (Yuyama et al., 2011; Grigio et al., 2021).
Due to the importance of the species, the Brazilian Agricultural Research Corporation, in partnership with the Federal University of Roraima, has been conducting studies on prospection, selection, collection, cloning, and clonality testing with the purpose of domestication, conservation, and cultivation of the species in ecosystems other than their natural habitat. The search for quality genetic material has been challenging for researchers due to the significant genetic variability of wild populations caused by pollination, with 91% allogamy and 9% autogamy, giving rise to extensive heterogeneity in the agronomic characteristics of interest in production (Vásquez, 2000).
Recommendations suggest working with an adequate number of genetic materials to avoid the reduction of variability that may lead to future problems of vulnerability to pests and diseases. Assessing the plant initial developmental characteristics allows us to determine the growth efficiency and the plant ability to adapt to environmental conditions. However, in most cases, selection based on one or a few characteristics may be inadequate since it must be carried out with superior products selected using several morphological characters (Costa et al., 2005; Faleiro & Junqueira, 2011). Thus, the isolated evaluation of growth characters cannot be accepted for assigning a quality index or merit selection, confirmation, or recommendation of quality clones.
Similarly, according to Ventura et al. (2012), it is difficult to define which characteristics should be evaluated for the improvement programs. Therefore, the authors suggest using multivariate analysis techniques to determine possible correlations and, thus, identify those responsible for most of the total variation observed. These techniques are useful for evaluating individuals in several aspects, providing a holistic view of each genotype (Cruz et al., 2012).
Another technique used is grouping analysis that aims to gather and classify the genotypes in several groups, following some similarity or dissimilarity criteria based on morphological and agronomic characters (Araújo et al., 2002; Alves et al., 2003; Martel et al., 2003).
The objective of this research was to select camu-camu clones with superior initial development using multivariate techniques at two years of planting under savanna/forest transition conditions of Roraima, northern Amazon.
Materials and methods
The research was conducted at the "Serra da Prata" Experimental Field of the Brazilian Agricultural Research Company, Embrapa-RR, located in the municipality of Mucajaí, at 60°58'40" W and 2°23'49" N, representative of the savanna/forest transition area of the state of Roraima. Historically, the site had been fallowed for four years, with a monthly clearing of spontaneous vegetation.
According to the Köppen classification system, the climate is Am type, with a short dry season. The longest rainy season is from May to July, while the shortest is from October to March, with an annual average of 1,844 mm (Mourão et al., 2003).
We used 56 camu-camu clones from 11 native populations characterized and selected from December 2014 to April 2015 by Chagas et al. (2015). The clones were produced by rooting piles in sub-irrigation chambers, following the method described by Abanto et al. (2014). After 12 months, when the clones had an average height of 80 cm, they were transplanted to the experimental field (June 2016).
The clones used were: 1) AB-04; 2) AB-05; 3) AB-06; 4) AB-07; 5) AB-08; 6) ABU-01; 7) ABU-02; 8) AT-03; 9) AT-07; 10) AT-08; 11) AT-10; 12) AT-13; 13) BQ-03; 14) BQ-04; 15) BQ-12; 16) BQ-18; 17) BQ-26; 18) BQ-27; 19) BQ-28; 20) BQ-29; 21) BQ-32; 22) EV-06; 23) EV-07; 24) EV-09; 25) EV-10; 26) IAB-01; 27) IAB-02; 28) IAB-04; 29) IAB-05; 30) IAB-06; 31) IAB-07; 32) LM-08; 33) LM-26; 34) LM-27; 35) LM-29; 36) LM-30; 37) LM-31; 38) LM-39; 39) LM-47; 40) LR-03; 41) LR-04; 42) LR-05; 43) LR-11; 44) LR-12; 45) MU-06; 46) MU-11; 47) MU-16; 48) MU-17; 49) RQ-04; 50) RQ-11; 51) RQ-14; 52) RUR-01; 53) RUR-02; 54) RUR-03; 55) RUR-04, and 56) RUR-05.
The area chosen for the clonality test was 5,040 m2. Before the crop was established, the area was cleared, plowed, and gridded so that all the vegetal biomass remaining in the superficial layer of the soil was incorporated. The level of soil fertility was very low, with a low sum ofbasic exchangeable cations (S) ranging from 0.1 to 0.8 cmolc kg-1 of soil; the cation exchange capacity (CEC) was lower than 2.1 cmolc kg-1 of soil; the low base saturation ranged from 7% to 46%, and the aluminum saturation was greater than 50% in most profiles (Rodrigues et al., 2000). Therefore, 1,600 kg ha4 of dolomitic limestone was applied of which 1,550 kg ha-1 was applied to the set, and 50 kg ha-1 was applied to the pits (150 g/plant). The pits were opened with dimensions of 0.40 m x 0.40 m and received 150 g dolomitic limestone 30 d before transplanting. At the time of transplanting, 67 g of urea, 110 g of Super Triple fertilizer, 103 g of KCl, and 10 g of slow release micronutrient (FTE BR-12®, Nutriplant, Brazil) were applied per pit. In the second year of cultivation, 111 g of urea, 122 g of Super Triple, and 121 g of KCl were applied.
Drip irrigation was used; the emitters had a flow rate of 3.4 L h-1 with a 50 cm spacing. A Rain Bird® controller (Rain Bird, Azusa, CA, USA) automatically activated the irrigation system. Weed control was performed using a brushcutter, manually in the planting rows and attached to a tractor between rows.
The camu-camu clones were transplanted at 5 m x 3 m spacing (5 m between rows and 3 m between plants), arranged in a random block delineation with three blocks and two plants per experimental unit. The evaluation of the vegetative characteristics was carried out 12 months after planting.
The variables measured were as follow: plant height [H (m)], number of basal branches (NBB), number of terminal buds (NTB), basal stem diameter [BSD (mm)], contents of chlorophyll a (Chl a) and b (Chl b), and total chlorophyll (Chl a + b). Chlorophyll a and b were determined with the Falker clofiLOG® chlorophyllometer. The measurement was made on 6 leaves from the middle part of the main branch of all the clones; the measurement was not performed on the leaves of the apical and basal part of the branch.
Principal component analysis was performed (PCA) and conglomerate (cluster) analyses based on Mahalanobis genetic distance (D2), both performed using Infostat software (Di-Rienzo, 2008). Additionally, the vegetative quality index (VQI) of the camu-camu clones was determined using the joint analysis of H, NTB, BSD, and Chl a. The reference parameters were adopted from those reported by Abanto et al. (2011) and Abanto-Rodríguez et al. (2016). For H, 2.96 m was used as a reference; for NTB, a value of 70 was used; 30 mm for BSD; and 37.15 for Chl a content. The analysis of VQI was an adaptation of the methods proposed by Karlen and Stott (1994)), Islam and Weil (2000), Melo Filho et al. (2007), and Freitas et al. (2012) for the determination of the soil quality index (SQI).
VQI consists of the differences between the evaluated attributes at initial development compared with the baseline of the quality reference attributes existing in the literature that were then calculated and expressed as the means of the individual value deviations for each attribute. The overall mean of each vegetative development attribute's deviations represents its deterioration from the reference (Freitas et al., 2012).
VQI was calculated in two steps, according to equations 1 and 2,
Where Qv refers to the mean of the deviation of the indicators of each attribute from the reference attributes present in literature; w refers to the value of the indicator measured in the systems under study; k refers to the value of the indicator measured in the reference system; n is the number of indicators that compose each set of attributes that in this study is 4; and Qvq is the mean of the deviations of the morphological characteristics of plants.
Results and discussion
Principal component analysis (PCA)
For the PCA, we initially considered the seven characters evaluated for the 56 camu-camu clones. However, after the first analysis, we found that PC1 and PC2 represented 74% of the data total variability. Therefore, seeking to improve the value of variability according to the criteria of Cliff (1998), we chose to discard variables following the methods described by Jolliffe (1972) and Mardia et al. (1979).
The criterion adopted for character discrimination was based on the correlation input (Mardia et al., 1979), where characters with minimal correlation are discarded. Thus, the NBB variable was discarded since it was minimally correlated with the other variables and principal components CP1 and CP2 (Tab. 1).
TABLE 1 Correlation matrix of original core variables and principal components of vegetative development of clones from different populations of transplanted camu-camu in the savanna/forest area of the northern Amazon, Brazil, 2018.

Cophenetic correlation=0.92.
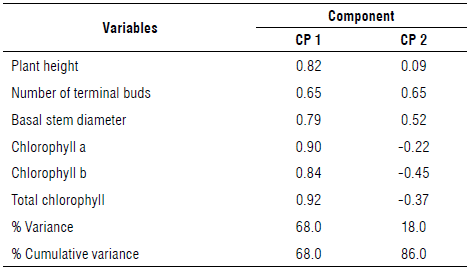
After discarding NBB, a new PCA was performed. Table 2 explains the total variance for all the principal components generated. The variance associated with each principal component was different and decreasing. The first principal component (PC1) accounted for 68.0%, while the second component (PC2) accounted for 18.2% of the total variance, with a persistent downward trend until all the variability was distributed among the six principal components generated. Thus, CP1 and CP2 represented 86.2% of the total data variability.
TABLE 2 Core values and proportion of variance explained by PCA analysis of vegetative development characters of camu-camu clones of different populations.

The distribution of coefficients in the principal components (Tab. 3) indicated that H, NTB, BSD, Chl a and b, and Chl a + b had the most positive contribution to CP1 with adequate correlation levels; therefore, these variables were the most responsive to camu-camu clone selection. However, the variables contributing positively to the principal component 2 (PC2) were NTB and BSD.
TABLE 3 Correlation matrix of the original core variables and principal components of vegetative development of camu-camu clones from different populations transplanted for clonality testing.
Cophenetic correlation=0.923.
Figure 1 shows a biplot with the two principal components explaining a significant proportion of the variability of the 56 camu-camu clones recorded data. The clones were scattered, demonstrating variability, although some were of the same origin. The first principal component (PC1) contributed 68.0% of the total variance while the second component (PC2) accounted for 18.2%, bringing the total variability to 86.2%, an acceptable value given that it surpasses the 70% threshold put forward in the criteria of Cliff (1998).

FIGURE 1 Distribution of clone variables from different populations of transplanted Myrciaria dubia in a clonality trial in the transitional savanna/ forest area of the northern Amazon. Plant height (H), number of terminal buds (NTB), basal stem diameter (BSD), chlorophylls a (Chl a) and b (Chl b), and total chlorophyll (Chi a + b).
PC1 associated the clones of the Branco River populations [Água Boinha (AB-07, AB-06), Lago da Morena (LM-47, LM-29), Lago Muçum (MU-17), Bem Querer (BQ-04, BQ-18, BQ-26, BQ-29, BQ-28), Açaí Tuba (AT-10, AT-13, AT-03), Lago do Rei (LR-03, LR-11, LR-05) and Estirão do Veado (EV-09)); Mucajaí River [Igarapé Água Boa (IAB-01, IAB-02, IAB-07)], and Quitauaú River [Quitauaú (RQ-14)] with the variables that had the highest correlation coefficients.
We infer that all these clones presented desirable results since they positively correlated with all variables associated with PC1. Thus, the increase in the height of the camu-camu plant caused an increase in the BSD as well as a greater number of secondary branches, allowing significant development of leaf area and, consequently, greater exposure to light, photosynthetic activity, and vigor, reflected by the considerable content of Chl a, b and Chl a + b. Accordingly, there were camu-camu clones that showed a differentiated response to the application of nitrogen fertilization, reflected in the significant gains in Chl a and b and Chl a + b content.
The lack of N absorption and low Chl production by some camu-camu clones, possibly, led to inadequate utilization of sunlight for photoassimilate production that may have considerably limited development (Larcher, 2004). Similar observations were made by Colodetti et al. (2014), who reported that the significant genetic variability of coffee (Coffea canephora) allowed for the identification of genotypes with a greater capacity to use nutrients under different soil conditions.
The principal component PC2 explained 18.2% of the total variance (Fig. 1), where NTB and BSD had a significant positive contribution, associating with clones from Branco River [Lago da Morena (LM-26), Água Boinha (AB-04, AB-05), Lago Muçum (MU-11, MU-16), Lago da Morena (LM-39), Bem Querer (BQ-12), Açaí Tuba (AT-13)], and Mucajaí River [Igarapé Água Boa (IAB-05)].
Hierarchical cluster analysis
The cluster analysis of the 56 camu-camu clones (Fig. 2) distinguished 13 clone groups according to the dissimilarity of the vegetative characteristics assessed during the first two years after transplantation. In this analysis, the dendrogram's cut-off point was assumed to be a genetic dissimilarity of 1.85 (50%) among all the clones evaluated. Thus, using the six characteristics of initial development as dissimilarity patterns, we have the formation of two larger groups, groups G8 and G9.

FIGURE 2 Classification dendrogram based on the initial development characteristics of the 56 clones of the different Myrciaria dubia populations transplanted in a clonality test using the average linkage mean and Mahalanobis distance algorithm (Cophenetic correlation=0.702).
The largest group (G9) comprised 20 clones (from LM-27 to AB-05, not in sequential order), that is, 35.7% of all clones that came from the Branco River [Lago da Morena (LM-07, LM-08, LM-30, and LM-39), Bem Querer (BQ-03, BQ-12, and BQ-27), Estirão Veado (EV-06 and EV-10), Lago Muçum (MU-06), Açaí Tuba (AT-03, AT-07, and AT-08), Água Boinha (AB-05 and AB-06), Lago do Rei (LR-04)]; Mucajai River [Igarapé Água Boa (IAB-04 and IAB-06)], and Urubù River [Urubù (RUR-03 and RUR-04)].
The second group (G8) consisted of 15 clones (from RQ-11 to AB-06, not in sequential order), representing 26.8% of the clones evaluated, coming from the following rivers and populations: Quiatuaú River [Quiatuaú (RQ-11 and RQ-14)], Urubù River [Urubù (RUR-01)], Unimirim River [Água Boa (ABU-02)], and Branco River [Bem Querer (BQ-04), BQ-18, BQ-26, BQ-28, and BQ-29), Good Water (AB-07 and AB-06), Lago do Rei (LR-05 and LR-11), Lago da Morena (LM-31), and Lago Muçum (MU-17). This group was formed by eleven clones of the Branco River population, two clones of the Quitauaú River, and one clone of the Unimirim and Urubú Rivers.
Thus, the G8 and G9 contained 35 of the 56 clones (62.5%). The larger groups represent materials whose dissimilarity could not be identified. Group 12 (Gl 2) consisted of six clones from the Branco River and the following populations: Lago Muçum (MU-11 and MU-16), Lago da Morena (LM-29 and LM-47), Lago do Rei (LR-03), and Açaí Tuba (AT-13)). Group 6 (G6) was formed by three clones from the following rivers and populations: Mucajai River [Igarapé Água Boa (IAB-01 and IAB-02)], and Branco River [Açaí tuba (AT-10)]. It should be noted that this group had two clones from the Mucajai River and one from the Branco River. Group 3 (G3) was formed by two clones from the rivers and populations of the Urubù [Urubù (RUR-04)] and Branco Rivers [Lago da Morena do Rei (LR-12)]. Group 4 (G4) was formed by clones from the Urubù [Urubù (RUR-05)] and Unimirim [Agua Boa (ABU-01)] Rivers, and Group 13 (G13) by clones from the Branco [Agua Boinha (AB-04)] and Mucajai [Igarapé Água Boa (IAB-05)] Rivers.
Finally, groups Gl, G2, G5, G7, G10, and Gil were formed in isolation by a clone each from the rivers and populations of the Branco River [Estirão do veado (EV-07)]; Quitauaú River [Quitauaú (RQ-04)]; Branco River [Estirão do veado (EV-09)]; Branco River [Bem Querer (BQ-32)]; Mucajai River [Igarapé Água Boa (I AB-07)] and Branco River [Lago da Morena (LM-26)], respectively.
These results confirm the existence of variability in the genotypes evaluated based on all the characteristics evaluated together. Similar results were obtained by Yokomizo et al. (2017) when determining the genetic diversity among the progeny of açai plants from Anajás, Pará, Brazil.
This diversity collection occurs due to obtaining materials from native populations (Yokomizo et al., 2017). Thus, the presence of genetic variability could be explored in genetic improvement programs to obtain superior genetic combinations and eventually be used in future controlled crossings avoiding the use of clones belonging to the same group.
Souza et al. (2010) reported that estimating genetic distances between clones is essential; from our results, it is possible to suggest crosses or follow up potential crosses to explore hybrid vigor between progenies. Souza et al. (2010) and Cruz et al. (2011) highlight that the dissimilarity analysis (distance) allows evaluating possible redundancies in a set of individuals. However, crosses between more distant individuals make it possible to uncover greater segregation in future improvement cycles and, therefore, enrich their genetic basis.
The cophenetic correlation in the present study was 0.702, indicating that the groups formed are reliable. Alvarado et al. (2016) obtains similar results, working with the agronomic characterization of 95 coffee accesses, and Santos et al. (2014), reports a 0.69 cophenetic correlation in a group analysis of cupuaçu plant clones. Cruz et al. (2011) indicate that the estimated coefficients range from zero to one, and the higher the value obtained for this coefficient, the higher the representativity of the dendrogram in relation to the genetic distance matrix.
Vegetative quality indices (VQI)
Figure 3 shows the clones with the highest VQI, resulting from the joint analysis of the initial development variables evaluated in the first two years of age. In this type of analysis, the position in order of merit can be determined more precisely according to all the evaluated attributes. Thus, clones with higher VQI were from the following rivers and populations in descending order: Branco River [Água Boinha (AB-05)]; Mucajai River [Igarapé Água Boa (IAB-05)]; Branco River [Lago Muçum (MU-11), Bem Querer (BQ-04), Lago da Morena (LM-26), Açaí Tuba (AT-10), Estirão do Veado (EV-06), Lago do REI (LR-04), Água Boinha (AB-06), Lago da Morena (LM-29), Água Boinha (AB-04), and Açaí Tuba [(AT-13), (AT03)]; Lago da Morena (LM-39); Lago do Rei [(LR-03, LR-11, and LR-05)]; Lago Muçum (MU16); Bem Querer (BQ-12); Mucajai River [Igarapé Água Boa (IAB-06)]; Branco River [Bem Querer (BQ-29)]; Lago da Morena (LM-47)]. The clones with the smallest VQI were: RUR-05, RUR-02 and ABU-01, belonging to the rivers and populations of the Urubù (Urubù) and Unimirim Rivers (Água Boa).
Conclusions
Plant height, number of terminal buds, basal stem diameter, chlorophylls a and b, and total chlorophyll contributed the most to the camu-camu clone selection in the savanna/forest area of the northern Amazon, Brazil. We verified that 22 clones of camu-camu were better adapted to soil conditions out of the floodplain. In this sense, this study represents a great advance for the domestication of the species, since there are more clones available to be included in the camu-camu genetic improvement program of the Brazilian Agricultural Research Corporation-EMBRAPA Roraima.
















