Introduction
Coffee fermentation is a crucial step in the production of high-quality coffee, and the microorganisms involved in the process play a significant role in determining the flavor and aroma of the final product 1,2. The unique growing conditions and fermentation methods used in Colombia coffee production have contributed to the country's reputation establishing it as the third major coffee-producing country 3,4. However, there have been some challenges in the post-harvest practices, particularly in the fermentation process, leading to occasional defects and inconsistencies in quality. These issues not only affect the coffee's overall quality but also result in economic losses for the farmers 5.
The fermentation of coffee fruits aims to degrade the mucilage by indigenous bacteria that feed on various components of the pulp and mucilage. Microbial diversity depends on many factors, including the agricultural area, coffee variety, harvesting, and coffee cherry storage 6,7. Producers and some researchers only indicate fermentation for mucilage removal but do not evaluate the impact on aroma and flavor. In fermented green coffee, mucilage removal must be effective and contribute to the quality due to the biochemical changes resulting from their degradation. Besides the impact on the beverage quality, there are fermentation benefits such as decreased fungi contamination 8.
During fermentation, there are physicochemical changes in the grains, such as a reduction in the water and simple sugars content and production of organic acids, alcohols, enzymes (poly-galacturanase, pectinlyase), and other metabolites. All these compounds are precursors of the coffee bean’s aroma and flavor, influencing the beverage’s final sensory characteristics 5,9. Wet fermentation is one of Colombia’s most widely used methods for coffee fermentation. In this method, the skin and pulp are removed mechanically, leaving some of the mucilage adhered to the beans. These de-pulped coffees are then transferred to water tanks, where they ferment for 6 to 72 hours, depending on the environmental temperature. The remaining mucilage is degraded and solubilized. The beans are then removed from the tanks and dried 9.
For over a century, the wet method has been a water-intensive process, as the name indicates, and there was little concern for water consumption and contamination 10. The constant market demand for high-quality coffees and sustainable production processes has prompted the exploration of the environment and microbiological interactions in wet coffee fermentation. This research aimed to study cultivable and viable microbial communities in the coffee from the wet fermentation process of Coffea arabica varieties Catimor and Castillo found at Finca La Antigua (Jardín, Antioquia, Colombia). Results generated from this study will allow knowing the diversity of viable microorganisms present in coffee fermentation under natural environmental conditions and the variables studied. These microorganisms can be” starter cultures” used in controlled fermentation processes to produce Specialty Coffees in Colombia.
Materials and methods
Location and coffee treatment
Coffee cherries from “La Antigua'' farm (5.608263510940495, -75.81934403607546) located at 1,850 m above sea level in Jardín, Antioquia, Colombia, were hand-picked at the mature stage (cherries) and then were classified (red cherries) comparing the cherries with a color chart (color codes: #fc4f59, #ef2b2d, #d62828, #af2626, #ef2b2d, #cc2d30, #a03033, #ce1126, #af1e2d) according to reference 11. They were de-pulped in a traditional pulper (Vencedora Estrella no. 3 1/2) to obtain beans with mucilage. The pH (UdeA device, Colombia), were measured from the fresh de-pulped coffee beans and ºBrix (Ultechnovo, USA) were measured from the cherries, by squeezing a drop of juice from cherries into a hand refractometer.
Coffee fermentation conditions
Our experimental unit was a mini-batch (plastic containers without lids (13x30x20 cm)). We used nine mini-batches with 18 Kg of de-pulped coffee cherries (a mixture of C. arabica var. Catimor and Castillo). Each one had a different water (g)/de-pulped coffee (g) ratio (I: 0/25, II: 10/25, III: 20/25) and a final time of fermentation (A: 24h, B:48h, C:72h). The pH was registered through the fermentation process of the pulped coffee beans. ºBrix was measured at the end of each fermentation experiment. The experimental design consisted of a three-by-two factorial design, being the factors the water (g)/pulped coffee (g) ratio and the fermentation time. Each experiment was done by duplicate. Each experiment was done by duplicate.
Microbial count and isolation of lactic-acid bacterial and yeast
300 g of samples from each mini-batch were collected in sterile plastic bags in three moments of fermentation: t0 (beginning), t2 (half-time:12h, 24h, 36h), and t3 (final time: 24h, 48, 72h). Afterward, each sample was stored at 4ºC and then transported in ice boxes to the Food Microbiology laboratory at Universidad de Antioquia, Medellín, Colombia. 10 g of each sample was added to a bottle containing 90 ml of saline-peptone water (0.1% bacteriological peptone, Merck, USA). After mixing, six-fold dilutions were prepared. Microorganisms count was carried out using five different culture media and plating in duplicate using 50 μl of dilutions 10-4 and 10-6 as follows: mesophiles microorganisms in Plate count agar (PCA, Microkit, Spain) and incubated at 32 ºC for 72 h; acetic-acid bacteria (AAB) in Wallerstein Lab nutrient agar (WL, Scharlau, Spain) and incubated at 37 ºC for 24 h; coliforms in Chromocult (Merck, USA) and incubated at 37 ºC for 24 h; lactic-acid bacteria (LAB) in Man, Rogosa y Sharpe agar (MRS, Merck, USA) supplemented with Fluconazole 0.2 % p/v (La santé, Colombia); yeasts and filamentous fungi in Oxytetracycline-Glucose Yeast Extract agar (OGYE, Oxoid, UK) supplemented with 80 mg gentamycin (Genfar, Colombia) in 500 ml and incubated at 25 ºC for 72 h.
All colonies were counted and results were expressed in log CFU/g (Colony Forming Units per gram) and described by shape, color, height, and edge of each isolate. Gram staining and catalase tests were made. Yeasts and lactic-acid bacteria colonies with different macroscopic and microscopic characteristics were isolated, purified, and stored in vials at -80 ºC in 20 % glycerol.
Molecular identification of acid-lactic bacteria and yeast.
Molecular identification of lactic-acid bacteria (LAB) and yeasts colonies isolates was performed by MACROGEN (Korea) 12. The samples were sent according to MACROGEN’s (Korea) preparation guidelines 12. The amplification and sequencing of the 16S ribosomal gene were done using the universal primers: 785F (3' GGA TTA GAT CCC TGG TA 5') and 907R (5' CCG TCA ATT CCT TTR AGT TT 3'). Yeast molecular identification was performed by amplifying and sequencing of 26S rDNA region using the universal primers ITS4 (TCC TCC GCT TAT TGA TAT GC) and ITS5 (GGA AGT AAA AGT CGT AAC AA GG) for sequencing of 18S rDNA region. The microbial sequencing chromatograms were depurated using Benchling 13. The sequences were compared with yeasts and LAB references from the NCBI GenBank using the Basic Local Alignment Search Tool (BLAST) algorithms 14. Finally, phylogenetic trees were made using MEGA X software 15.
Data analysis
The experiment design was completely randomized in a factorial arrangement of three different fermentation times and three water(g)/de-pulped coffee(g) ratios for 9 treatments with 2 replicates. Analysis of variance (ANOVA) and Pearson correlation were conducted using R software 16. P-values less than 0.05 were considered statistically significant.
Results
pH and ºBrix in the coffee fermentation process.
pH and ºBrix in de-pulped coffee during the fermentation process are reported in Figure 1 and Table 1.
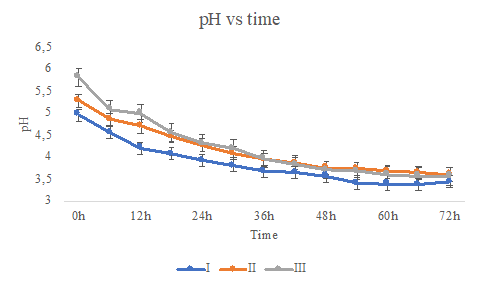
Figure 1 pH during fermentation of different water/pulped coffee ratios (I: 0/25, II: 10/25, III: 20/25) and final time of fermentation (A: 24h, B:48h, C:72h).
The water(g)/de-pulped coffee(g) ratio influenced the initial pH values of the mini-batch systems. A low positive correlation of 0.30 (data not shown) suggests a basic state in rich water media (mini-batch III: 5.82, mini-batch II: 5.28, and mini-batch I: 4.96). Additionally, the pH values in each system gradually decreased during fermentation and converged at the end of each process (mini-batch III: 3.56, mini-batch II: 3.56, and mini-batch I: 3.52).
The value of °Brix for all mini-batch treatments is presented in table 1. ºBrix of mature coffee cherries were in the optimal range of 15 and 15.8, corresponding to the optimum maturity of the coffee fruit Colombia variety 17. Mini-batches A and C started with slightly higher values but in general, the data shows a decreasing behavior of the concentration of mucilage sugars in all treatments.
Microbiological counts and isolation of lactic-acid bacteria and yeasts.
Bacterial and yeast populations in mini-batch fermentations were quantified by plating on selective media. Microbial counts of mesophilic aerobic bacteria, coliforms, AAB, LAB, and yeast found during coffee processing are illustrated in Figure 2 to Figure 6.
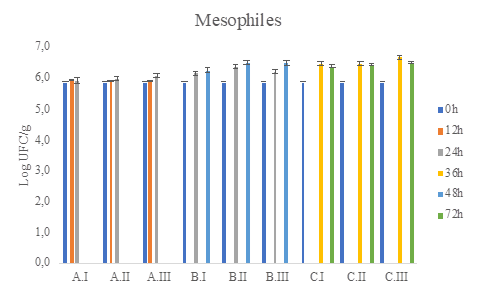
Figure 2 Total count (CFU / g) of mesophilic microorganisms in Plate count agar (PCA) medium during coffee fermentation (final time A: 24h, B:48h, C:72h) and different water(g)/pulped coffee (g) ratios (I: 0/25, II: 10/25, III: 20/25).
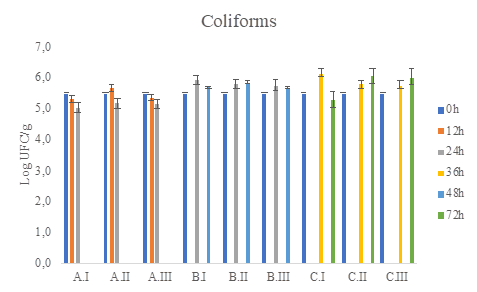
Figure 3 Coliforms total count (CFU / g) in Chromocult medium (CHC) during coffee fermentation (final time A: 24h, B:48h, C:72h) and different water(g)/pulped coffee(g) ratios (I: 0/25, II: 10/25, III: 20/25)
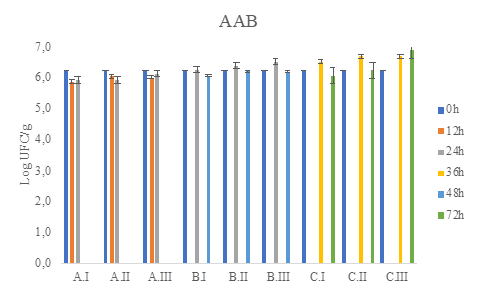
Figure 4 Acetic-acid bacteria total count (CFU / g) of in WL medium during coffee fermentation (final time A: 24h, B:48h, C:72h), and different water/pulped coffee ratios (I: 0/25, II: 10/25, III: 20/25).
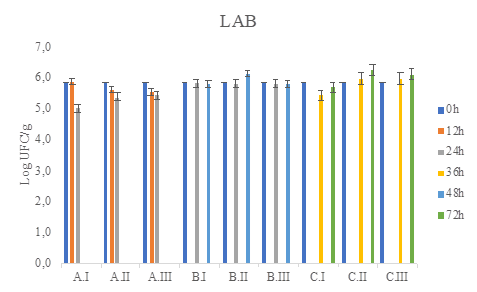
Figure 5 LAB total count (CFU / g) in MRS medium during coffee fermentation (final time A: 24h, B:48h, C:72h), and different water/pulped coffee ratios (I: 0/25, II: 10/25, III: 20/25).
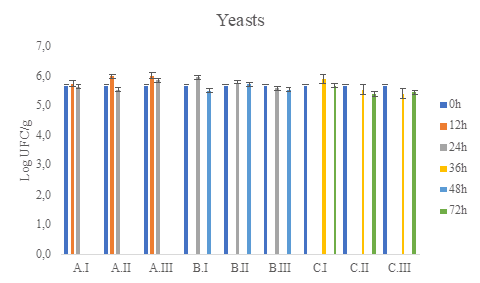
Figure 6 Yeasts total count (CFU/ g) in OGYE medium during coffee fermentation (final time A: 24h, B:48h, C:72h), and different water/pulped coffee ratios (I: 0/25, II: 10/25, III: 20/25).
Table 2 presents the statistical analysis data (P-values) of the effect of each factor and both factors combined (water/de-pulped coffee ratio and fermentation final time) over the counts in each microbial group.
Table 2 P- values obtained of statistical evaluation of water/de-pulped coffee ratio and final time fermentation factors over microbiological counts p- values.
| Water/de-pulped coffee | Time | Combined effect of factors* | |
|---|---|---|---|
| Mesophiles | 0.7084 | 0.1245 | 0.9809 |
| Coliforms | 0.415 | 0.308 | 0.744 |
| Yeasts | 0.821 | 0.128 | 0.513 |
| LAB | 0.1084 | 0.0213 a | 0.3712 |
| AAB | 0.0538 | 0.0176 a | 0.0933 |
a p-value significance level >= 0.05
* Effect of Water/de-pulped coffee and Time on microbial counts
All bacterial and yeast isolates were identified according to 16S rRNA and ITS gene sequencing, respectively. Taxonomic identification of the isolates was determined by aligning LAB and yeast sequences and the species with a 99-100% identity percentage 18 were selected. The amount of species of lactic-acid bacteria and yeast isolates present in coffee fermentation are shown in Figure 7 and Figure 8. The percentages are the quantity of successfully recovered isolates, meaning viable and cultivable microorganisms.
The isolation of different lactic-acid bacteria (LAB) species corresponds to the first 48 hours of fermentation mini-batches A and B and water/pulped coffee ratios of I: 0/25 and II: 10/25. The most abundant LAB isolates correspond to Lactiplantibacillus plantarum (46%), and Levilactobacillus brevis (31%). Lactiplantibacillus pentosus, Lacticaseibacillus rhamnosus and Lactobacillus sp. had approximately the same recuperation; 8% for the first two species and 7% for Lactobacillus
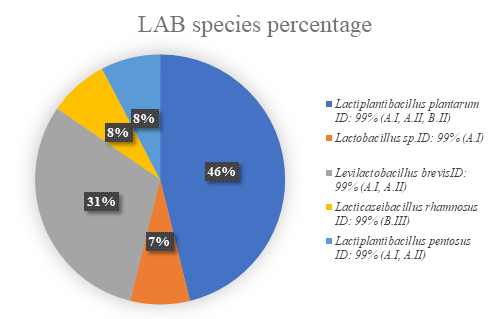
Figure 7 Species of lactic-acid bacteria present in coffee fermentation at final time (A: 24h, B:48h, C:72h), and different water/de-pulped coffee ratios (I: 0/25, II: 10/25, III: 20/25).

Figure 8 Species of yeast present in coffee fermentation at final time (A: 24h, B:48h, C:72h), and different water/de-pulped coffee ratios (I: 0/25, II: 10/25, III: 20/25).
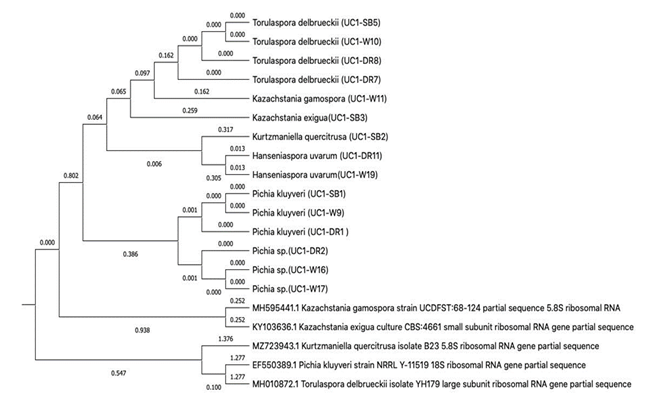
Genetic relationships between representative isolates of lactic-acid bacteria and yeasts in coffee fermentation are represented in phylogenetic trees (Figures 9 and 10). The phylogenetic trees were built to establish the evolutionary distance within each group of LAB and yeasts identified in this study and primally assess if they could be safe to be used as starter cultures in other fermentations or if they have been reported as species generally recognized as safe (GRAS) in the National Center for Biotechnology Information (NCBI) database.

Figure 9 Phylogenetic relationship between the identified lactic acid bacteria from wet coffee fermentation and other 16S rRNA sequences of published strains.
Discussion
pH in coffee fermentation process.
According to the results shown in figure 1, the water(g)/de-pulped coffee(g) ratio influenced the initial pH values of the mini-batch systems. The decrease in the pH during coffee bean fermentation has been reported as the degradation of mucilage’s components (e.g., pectic substances, complex sugars, amino acids, and proteins) 19 into simpler sugars due to the action of microorganisms 20.
ºBrix in the coffee fermentation process.
The standard deviation (SD) shows a low variability of the ºBrix throughout time (Table 1). Regardless of the water (g)/de-pulped coffee (g) ratio used in the mini-batch fermentation, the ºBrix decreased at the end of all the experiments since microorganisms consumed sugars to develop their metabolic processes. The aforementioned is in agreement with other studies 8. For example, Pantoja López.21 reported a ºBrix exponential decline in a study conducted in Huila, Colombia, and Betancur Henao.22 obtained the same results in southwest Antioquia, Colombia.
Microbiological counts and isolates of lactic-acid bacteria and yeasts.
In table 2 is observed that the fermentation time factor had influence over the LAB and AAB counts, but the combination of the factors; time and water/de-pulped coffee level ratio, did not show there was a significant amount difference in microbial counts (P-value > 0.05).
The viable and cultivable counts of the different microbial groups analyzed in the wet fermentation were between 105 and 106 log CFU/g of fermented coffee (Figure 2 to Figure 6). As no treatment exceeds at least 3 logarithms above or below the counts it is concluded that there are no statistically significant differences
LAB has been recognized as an integral component of coffee processing in most coffee-producing countries 23-25. The LAB growth is because of adaptability to the environment and stress factors of coffee processing, such as pH variation, sugar availability, and competition with other microorganisms 23,26,27. LAB species can catabolize pentoses and hexoses in coffee pulp into a vast range of end-metabolites, including lactate, acetate, CO2, and ethanol, via the phosphoketolase or pentose phosphate pathway 28.
The results revealed that the most prevalent yeast species were Pichia kluivery, accounting for 39% of the isolates, followed by Torulaspora delbrueckii at 22%. Hanseniaspora uvarum, Kurtzmaniella quercistrusa, and Kazachstania gamospora were also detected, although at lower amounts of 11%, 5% and 6%, respectively.
The presence of Pichia kluivery, Torulaspora delbrueckii and Hanseniaspora uvarum, as the dominant yeast isolates suggests their potential role in driving the fermentation process and influencing the final coffee product. The use of these yeast species as starter cultures has been studied due to their association with desirable sensory characteristics and are recognized for their ability to contribute to the development of distinct flavors and aromas, thereby playing a crucial role in the production of specialty coffee 2,29,30.
Lactic acid bacteria were isolated from the first 48 hours while yeasts were isolated after 48 and up to 72 hours. LAB are well-adapted to the pre-existing environmental conditions; therefore, it is common to find LAB species instead of yeasts in the initial hours of coffee fermentation 25,31. The microbial community is known to vary in composition and initial abundance during this process, and the succession of bacterial and fungal species is complex because their behavior depends on the time and environmental factors (32-34.
A phylogenetic tree analysis was conducted to examine the genetic relationships among representative isolates of lactic acid bacteria (LAB) species belonging to the Lacticaseibacillus, Lactiplantibacillus, and Levilactobacillus genera (Figure 9)
The phylogenetic tree revealed distinct clusters representing each genus. The Lacticaseibacillus and Lactiplantibacillus cluster comprised species within the plantarum group lactobacilli. These clusters showed a close evolutionary relationship to other lactobacillus groups commonly used as starter cultures in fermentations 35, antimicrobials 36,37, and the next generation of probiotics (38,39. This suggests a shared genetic heritage and functional similarities among these lactobacilli.
The Levilactobacillus cluster represented a group of lactobacilli known for their leavening potential 40. Multiple species within this genus have been identified in type sourdoughs used as leavening agents. The presence of Levilactobacillus highlights the diversity and adaptability of these lactobacilli in different fermentation environments (40.
Understanding the genetic relationships among these species contributes to our knowledge of their diversity, evolutionary history, metabolic capabilities, and potential applications. It provides a framework for further investigation into the functional properties, ecological roles, and their impact on fermentation and product quality.
The yeast isolates identified in this research belong to five different genera (Figures 8 and 10), namely Torulaspora, Kazachstania, Kurtzmaniella, Hanseniaspora, and Pichia. Of particular interest is the isolation of Torulaspora delbrueckii, which has recently been reported in metagenomic studies conducted in Colombia 41 and has also been investigated in Latin America 42. This highlights the presence and potential significance of T. delbrueckii in coffee fermentation processes. Yeast species from the genus Kazachstania, including Kazachstania gamospora, have been documented in both Colombia 43 and Rwanda 44. This suggests that these yeasts are not limited to a specific geographic region and may have a broader distribution in coffee-growing areas.
Coffee processing steps have been found to harbor a diverse range of yeast species from various genera such as Pichia, Candida, Saccharomyces, and Torulaspora45. The presence of these yeast species broadens the understanding of the diversity and distribution of yeast species in coffee processing.
The analysis of LAB and yeasts in coffee fermentation provides valuable insights into the diversity and functional attributes of these microorganisms; including the production of flavor-active compounds and the establishment of desirable fermentation profiles. Further investigations into the metabolic pathways and interactions of these microorganisms will contribute to optimizing coffee fermentation processes and enhancing the quality of the final coffee product.
Conclusions
This research study aimed to know the influence of fermentation time and water on the development of viable microorganisms. Firstly, the study revealed that no treatment had an effect on the different microbial groups without controlling other environmental factors such as temperature and anaerobiosis. The identification of diverse bacterial and fungal species, including Pichia, Torulaspora, and Lactiplantibacillus, emphasizes their potential as starter cultures in controlled coffee fermentations and their relevance in fermentation technology and other biotechnological applications. The findings in this study are important for the future production of sustainable high-quality coffees and the development of starter cultures that have the potential of developing distinctive flavor profiles and aromatic compounds.