INTRODUCTION
The potential of an oocyte to develop into a viable embryo depends on the accumulation of mRNA or imprinted genes during the process of oogenesis. Thus there is a general agreement that the development competence of oocyte could be related to an abundance of a specific RNA transcript pool that is accumulated during oocyte growth and at final phases of folliculogenesis 1, favoring fertilization, implantation and generate healthy offspring. To clarify the importance of the contribution of the oocyte to the embryo quality, it is important to define more precisely the different types of competence expressed by oocytes. The ability to resume meiosis, to cleave upon fertilization to develop into a blastocyst, to induce pregnancy and to generate an healthy offspring are all separate events and succeeding in the first events does not ensure the success of subsequent ones. Furthermore, these events are associated with the three types of maturation processes observed in the oocyte: meiotic, cytoplasmic and molecular. These abilities vary also upon the type of follicle the oocytes is removed from. Larger or slow-growing follicles have been shown to foster better eggs than small or actively growing follicles. Hormonal stimulation can also affect oocyte competence with the nature of the effect (positive or negative). Folliculogenesis and follicle differentiation are processes that culminate in ovulation, being mainly coordinated by well-established endocrine mechanisms. This processes are the result of cellular and molecular transformations of the different structures that form the follicle 2.
Members of the transforming growth factor beta (TGFβ) super family and the heparin union growth factor family, are known to be important regulators of proliferation and differentiation of several types of cells 3. The growth differentiation factor 9 (GDF9), the bone morphogenetic protein 15 (BMP15), members of the family of growth transforming factor β (TGFβ), and the basic fibroblast growth factor (FGF2), member of the family of heparin union growth factors with their main receptors, have been involved in dominant follicle selection, regulating important events such as proliferation of granulosa cells, differentiation and steroidogenesis 4,5. GDF9 and BMP15 generate signals through their specific receptors, ie, transforming growth factor, beta receptor 1, (TGFβ-R1 or ALK5) and bone morphogenetic protein receptor, type IB, (BMPR-IB or ALK6), respectively, whereas FGF2 factor binds primarily to fibroblast growth factor receptor 2 (FGFR2) 6,7.
The expression of GDF9, BMP15 and FGF2 have been reported in rodents, sheep´s, buffalo and humans in oocyte, cumulus cells and ganulosa cells 4,5,8 and there is convincing evidence that they are important for ovarian function. However, there is little information about the expression of these factors in cows. Since cattle have been used for many purposes they are highly attractive livestock animals from an economic point of view. Therefore, it is desirable to improve the knowledge about their reproductive physiology, especially the control of ovarian folliculogenesis. In order to explore the intracellular modulatory system of GDF9, BMP15 and FGF2 in bovine ovaries, the goal of present study was to evaluate mRNA expression of GDF9, BMP15, FGF2 and their main receptors, TGFβ-R1, BMPR-IB and FGFR2, by using reverse transcriptase polymerase chain reaction (RT-PCR).
MATERIALS AND METHODS
Animals and follicular cells. Protocols used on animal were applied according to the standards and care described in the Official Journal of the European Union 9. Healthy bovine ovaries with normal morphology and no corpus luteum were collected from 70 ovaries obtained from 96 slaughtered beef heifers, during November-December 2015 and January-July 2016. The ovaries were placed into PBS solution at 37°C and delivered to the laboratory within 2 hours of collection. The cumulus-oocyte complexes (COCs) were recovered by manual aspiration of antral follicles (≤6 mm diameter) using 10 ml syringes with 40 x 1.2 mm (18 G) needles, and pooled in a sterile 50 mL conical bottom centrifuge tube with PBS. The tubes were kept at 37°C for 15 min for the settlement of COCs. The sediment was mixed with PBS and screened for COCs under a stereozoom microscope. The aspirated COCs were graded based on the morphological appearance of the cumulus cell investments and homogeneity of ooplasm as described by de Loos et al 10. Pools of 30 COCs qualitatively good (Grade 1 and Grade 2) were separated from the cumulus cells by repeated pipetting and then were recovered. Subsequently, three pools of 30 oocytes and three pools of cumulus cells, which were separated from the 30 oocytes, were made.
The follicular liquid, was centrifuged at 12.000 x g for 10 min at 4°C, obtaining three pellet of cells composed by granulosa cells and theca cells (PCs). All samples were snap-frozen in liquid Nitrogen and stored at -70°C until their analysis.
Extraction and purification of total RNA on follicular cells and oocytes. Total RNA was extracted from samples of follicular cells following the Phenol-Chloroform method using TriReagent® (Sigma) according to manufacturer instructions. Briefly, samples were homogenized with 250 µl TriReagent®, then 50 µl chloroform was added and merged for 15 sec followed by centrifugation at 12.000 x g for 15 min at 4°C. The upper aqueous phase, containing RNA, was transferred to a new tube and isopropanol was added. Samples were kept at room temperature for 10 min and centrifuged at 12.000 x g for 10 min at 4 ºC. Supernatant was removed and RNA pellet was washed with ethanol 75% and centrifuged at 7.500 x g for 5 min at 4°C. Total RNA samples were suspended on ribonucleases-free H2O and concentration and integrity of RNA were assessed by absorbance at 260 nm utilizing NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Scientific, U.S.A), and electrophoresis on Sybr Green stained agarose gels (Biotium). Prior to cDNA synthesis, DNase (AMPD1- Sigma) treatment was applied. Reverse transcription was performed using SuperScript III Reverse Transcriptase (Invitrogen). Every 20 μL of reaction contained 1 µl of Oligo dT (50 µM), 1µl of dNTPs, 4 µl of 5X First Strand Buffer, 1 µl of 0.1 M DTT, 40 units of RNAsin (Genbiotech), 200 units of SuperScriptIII reverse transcriptase, a volume of RNA (1.200 ng), and nuclease-free water to complete the final volume. The mixture was held 1 h at 50°C and 15 min at 70°C prior to cooling on ice. Finally, samples were stored at -70ºC till their subsequent analysis.
Primer design. Taking into account the sequences recently published by the National Center for Biotechnology Information (NCBI) and the data base of the bovine genome sequence, specific primers were designed in order to amplify bovine gene coding regions GDF9, BMP15, FGF2, TGFβ-R1, BMPR-IB and FGFR2 using the online software Primer-BLAST and Integrated DNA Technologies (IDT) (Table 1). Additionally, the quality and purity of samples used was determined with specific primers GAPDH (glyceraldehyde-3-phosphate dehydrogenase), for oocyte (ZAR1: zygote arrest-1), granulosa cells (CYP19A1: cytochrome P450, family 19, subfamily A, polypeptide 1) and theca cells (CYP17A1: cytochrome P450, family 17, subfamily A, polypeptide 1) as suggested Hatzirodos et al 11 (Table 1).
Table 1 Oligonucleotide sequences used for reverse transcriptase PCR assays (RT-PCR)
| Assay name 1 | Primers 2 |
|---|---|
| GDF9 | F: TGCACCTGTCTATGCCTTTG |
| R: AACATTTGGCCATGAGGAAG | |
| A: 157; AN: XM_010807219.1 | |
| BMP15 | F: TCAGGAAGAGGCTCCTCAAA |
| R: CCACCAGAACTCACGAACCT | |
| A: 168; AN: NM_001031752.1 | |
| FGF2 | F: AAGCGGCTGTACTGCAAGAA |
| R: GTAGTTTGATGTGTGGGTCGC | |
| A: 100; AN: NM_174056.4 | |
| TGFβ-R1 | F: GATTCGGCCACGGATACAA |
| R: GTCGAGCTACTTCCCAGAATAC | |
| A: 110; AN: NM_174621.2 | |
| BMPR-IB | F: TTTGGGAGGTCGCTAGGAGA |
| R: GCCGCAGCTTCTTGATACAC | |
| A: 132; AN: XM_005207720.2 | |
| FGFR2 | F: ATGTCGCTTATCGAGCCACC |
| R: GCGTGGCACCTTTTATCTGC | |
| A: 199; AN: XM_010820098.2 | |
| GAPDH | F:AGATGGTGAAGGTCGGAGTG |
| R:GAAGGTCAATGAAGGTCA | |
| A:117; AN: NM_001034034 | |
| CYP19A1 | F: GTGTCCGAAGTTGTGCCTATT |
| R: GGAACCTGCAGTGGGAAATGA | |
| A: 148; AN: NM_174305.1 | |
| CYP17A1 | F: ACCATCAGAGAAGTGCTCCGAA |
| R: CCACAACGTCTGTGCCTTTGT | |
| A: 115; AN: NM_174304.2 | |
| ZAR1 | F: TGCCGAACATGCCAGAAG |
| R: TCACAGGATAGGCGTTTGC | |
| A: 169; AN: NM_001076203 | |
| 1 Abbreviated mRNA transcript identifiers for GDF9: Growth differentiation factor, BMP15: Bone morphogenetic protein 15, FGF2: Fibroblast growth factor 2, TGFβ-R1: Transforming growth factor, beta receptor 1, BMPR-IB: Bone morphogenetic protein receptor, type 1B, FGFR2: Fibroblast growth factor receptor 2, CYP19A1: cytochrome P450, family 19, subfamily A, polypeptide 1, CYP17A1: cytochrome P450, family 17, ZAR1: zygote arrest-1 | 2 Details of specific primers sets used: F: forward primer sequence (5´to 3´), R: reverse primer sequence (5´to 3´), A: Amplicon length in base pairs and AN: Accession Number. |
Amplificaction by PCR. PCR reactions were performed by triplicate using cDNA of both follicular cells and oocytes. For this, mixtures containing 10 µM of each primer, 0.2 mM of each dNTP, 1 unit of Taq DNA polimerase (Genbiotech) and buffer solution of 10 X reaction (Tris-HCl 10 mM; KCl 50 mM; Triton X-100 0.1%; MgCl2 2 mM, pH: 9.0) in a 20 µl final volume. The amplification program used consisted of a denaturation step at 94°C for 5 min followed by 40 amplification cycles consisting of a denaturation step at 94°C for 20 sec, hybridization at 58°C for 30 sec, extension at 72°C for 1 min and finally a period of 7 min at 72°C. No-template and no-reverse transcription controls were included as negative controls and to exclude genomic DNA contamination. Specificity of PCR products was confirmed by electrophoresis on agarose gel and sequencing.
RESULTS
By using RT-PCR we demonstrated the presence of mRNA of GDF9, BMP15, FGF2, TGFβ-R1, BMPR-IB and FGFR2 on cumulus, follicular cell pellet (granulosa and theca cells) from antral follicles (≤6 mm diameter), as well as in oocytes. For all of mRNA studied we were able to obtain a single amplicon of expected size (Figure 1).
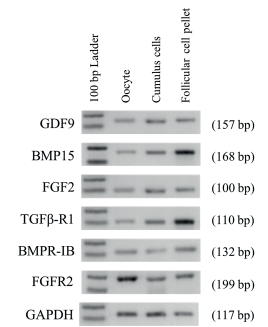
Figure 1 Expression of GDF9, BMP15, FGF2, TGFβ-RI, BMPR-IB, FGFR2 and GAPDH in different cell types in bovine ovaries. Cell types are indicated at the top (Oocyte, Cumulus cells, Follicular cell pellets). One hundred base pair ladders were included as markers for fragment size. Bands of 100 and 200 bp are shown in the 100 bp Ladder lane. Amplicon size are indicated on right in parenthesis.
In order to discard a possible cross contamination when each kind of cell was isolated, RT-PCR using specific primers was conducted. The mRNA expression of the CYP19A1 gene, specific for granulosa cells, only was detected in cumulus and follicular cell pellet; CYP17A1 gene, specific for theca cells, was detected only in follicular cell pellet. Finally, ZAR1 transcripts, specific for oocyte, were detected only in the cDNA template of the oocytes, but not in the cDNA template of cumulus cells, and follicular cell pellet.
DISCUSSION
Growth differentiation factor 9 plays multiple functions in oocyte and granulosa cell communication, regulation and differentiation 4. It has been suggested that GDF9 could influence follicular maturation and ovulation in humans by stimulating inhibin synthesis 12. The BMP15 gene also plays a critical role in ovarian function stimulating the proliferation of pre-antral granulosa cells and inhibiting FSH-stimulated progesterone production in later stage granulosa cells 4.
When comparing rodents with an incomplete estrous cycle (mice, rats) to animals with a complete estrous cycle (humans, goats, pigs and cows), GDF9 and BMP15 appear in different tissues. The mRNA expression of the transforming growth factors-beta studied was detected exclusively in oocytes of rodents 13, while in other species, including humans 14, goats 15, sheep 8,16, pigs 17 and cows 18,19, BMP15 and GDF9 were expressed in cumulus cells as well as in oocytes, in agreement with the result obtained in this work. Additionally, in the present study, BMP15 and GDF9 were also expressed in the pellet of cells, where granulosa and theca cells were expresed. These results could be explained by species specific differences in the mechanisms of action of key biological functions among the different follicle cells. In agreement with other authors, these differences could be explained by differences on physiological and reproductive forms of the studied species, suggesting that these factors regulate the ovulation rate and female fertility in a species-specific manner 20,21. Recently these species-specific functional differences between mono- and poly-ovulatory animals were attributed to the distinct timing of the transformation of the BMP15 pro-protein into a functionally mature BMP15 18.
GDF9 and BMP15 growth factors bind to specific receptors to trigger its mechanisms of action on the target cell. TGFβ-R1 mRNA expression was detected in granulosa cells of sheep and cows 19,22. BMPR-IB mRNA expression has been observed in goat, sheep, pig and bovine follicles 8,23,24, nevertheless here we report for the first time that TGFβ-R1 and BMPR-IB mRNA are expressed in bovine oocytes.
Fibroblast growth factors (FGFs) are implicated in pregnancy success by regulating embryogenesis, implantation, and placentation through cell survival, migration, and differentiation 25. FGF2, also known as basic FGF, has distinct roles during early embryogenesis in ruminants 26. FGF2 and its receptor FGFR2 are expressed during development of bovine embryos and a combination of FGF2 and transforming growth factor beta (TGFβ) improves the development of the bovine embryo 27. The FGF2 mRNA expression was detected in oocytes, cumulus granulosa cells, mural granulosa and theca cells strengthening the systemic role during folliculogenesis 7,8. It has been proposed that, oocyte-derived FGF2 could promote the primordial to primary follicle transition by signaling granulosa and stromal cells 28. In agreement, Khatib et al 29 suggest that FGF2 plays a positive role in the endogenous production of estrogens in humans 29. Furthermore, the angiogenic capacity of FGF2 described in follicles, generates proliferation of capillaries that follow the selection of the pre ovulatory follicle, resulting in a greater supply of nutrients and precursors, enhancing the growth of the dominant follicle 30. In previous work, it was suggested that FGF2 promotes mitosis of granulosa cells, theca, and ovarian stroma. It also enhances follicle survival, growth and formation of the antro during long term in vitro cultures of buffalo preovulatory follicles and improves steroidogenesis 30,31.
Although the biological significance of the expression of these factors in follicular somatic cells has not been fully elucidated, previous reports and the results of the present study indicate the possibility that these factors in somatic cells as well as in oocytes may regulate the selection of follicle and/or ovulation in species with a complete estrous cycle. The presence of GDF9, BMP15, FGF2, TGFβ-R1, BMPR-IB and FGFR2 transcripts in the studied cells indicate that these factors could act as paracrine and autocrine regulators in folliculogenesis.
Our results show the presence of a complex intrafollicular regulatory system, consisting of GDF9, BMP15, FGF2 and their main receptors in bovine ovaries, in a good agreement with previous studies on different species and follicular cells. Our results show an intra-follicular granulosa cell mRNA expression of the above genes from bovine ovarian follicles. These findings support the concept that early stages of ovarian follicular growth and development are regulated by intra-ovarian factors. Furthermore, our data showed that the initial concept that GDF9, BMP15 and FGF2 are exclusively produced by the oocyte cannot be maintained for the bovine species. To move forward with these findings, the quantitative levels of differential expression dynamics of GDF9, BMP15, FGF2, TGFβ-R1, BMPR-IB and FGFR2 in oocyte genes from bovine ovaries need further research.














