Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Universitas Scientiarum
Print version ISSN 0122-7483
Univ. Sci. vol.20 no.2 Bogotá May/Aug. 2015
https://doi.org/10.11144/Javeriana.SC20-2.mdas
Mitochondrial DNA analysis suggests a Chibchan migration into Colombia
Análisis mitocondrial de ADN sugiere urna migración Chibcha a Colombia
Análise de DNA mitocondrial sugere urna migração Chibchan à Colômbia
María Claudia Noguera-Santamaría1, 2, 5, Carl Edlund Anderson3, Daniel Uricoechea2, Clemencia Durán1, Ignacio Briceño-Balcázar1, 2, Jaime Bernal Villegas1, 4
Edited by Alberto Acosta & Juan Carlos Salcedo-Reyes
1Instituto de Genética Humana, Facultad de Medicina, Pontificia Universidad Javeriana. Bogotá, Colombia.
2Grupo de Genética Humana, Facultad de Medicina, Universidad de La Sabana. Chía, Colombia
3Department of Foreign Languages & Cultures, Universidad de La Sabana. Chía, Colombia
4Universidad Tecnológica de Bolívar. Cartagena, Colombia
5Facultad de Ciencias de la Salud. Grupo Gisafaco. Corporación Universitaria Remington. Medellín, Antioquia, Colombia.
Funding: Institute of Human Genetics — Pontificia Universidad Javeriana.
Electronic supplementary material: Suppl. 1-3
Received: 20-10-2014 Accepted: 01-07-2015 Published on line: 09-07-2015
Para citar este artículo / To cite this article
Noguera-Santamaría MC, Anderson CE, Uricoechea D, Durán C, Briceño-Balcázar I, Bernal-Villegas J (2015) Mitochondrial DNA analysis suggests a Chibchan migration into Colombia. Universitas Scientiarum 20(2): 261-278 doi: http://dx.doi.org/10.11144/Javeriana.SC20-2.mdas
Abstract
The characterization of mitochondrial DNA (mtDNA) allows the establishment of genetic structures and phylogenetic relationships in human populations, tracing lineages far back in time. We analysed samples of mtDNA from twenty (20) Native American populations (700 individuals) dispersed throughout Colombian territory. Samples were collected during 1989-1993 in the context of the program Expedición Humana ("Human Expedition'') and stored in the Biological Repository of the Institute of Human Genetics (IGH) at the Pontificia Universidad Javeriana (Bogotá, Colombia). Haplogroups were determined by analysis of RFLPs. Most frequent was haplogroup A, with 338 individuals (48.3%). Haplogroup A is also one of the most frequent haplogroups in Mesoamerica, and we interpret our finding as supporting models that propose Chibchan-speaking groups migrated to northern Colombia from Mesoamerica in prehistoric times. Haplogroup C was found in 199 individuals (28.4%), while less frequent were B and D, with 113 and 41 (16% and 6%) individuals, respectively. The haplogroups of nine (9) individuals (1.3%) could not be determined due to the low quality of the samples of DNA. Although all the sampled populations had genetic structures that fit broadly into the patterns that might be expected for contemporary Central and South American indigenous groups, it was found that haplogroups A and B were more frequent in northern Colombia, while haplogroups C and D were more frequent in southern and south-western Colombia.
Keywords: mtDNA; Native Americans; human migrations; Colombia; South America; haplogroups, RFLP Method - Restriction Fragment Length Polymorphism
Resumen
La caracterización del DNA mitocondrial (mtDNA) permite el estudio de estructuras genéticas y de relaciones filogenéticas en poblaciones humanas, al rastrear linajes hacia muy atrás en el tiempo. Analizamos muestras de mtDNA de veinte (20) poblaciones nativas americanas (700 individuos) dispersas por toda Colombia. Las muestras se obtuvieron entre 1989-1993 como parte del programa Expedición Humana y se almacenaron en el Banco Biológico del Instituto de Genética Humana (IGH) de la Pontifica Universidad Javeriana (Bogotá, Colombia). Los haplogrupos se determinaron mediante análisis de RFLPs. El más frecuente fue el haplogrupo A, con 338 individuos (48.3%). El haplogrupo A es también uno de los más frecuentes en Mesoamérica, y nosotros interpretamos nuestro hallazgo como una evidencia en favor de los modelos según los cuales los grupos que hablaban chibcha migraron al norte de Colombia desde Mesoamérica en tiempos prehistóricos. El haplogrupo C se encontró en 199 individuos (28.4%), mientras que los menos frecuentes fueron B y D con 113 y 41 individuos (16 y 6% respectivamente). No se pudieron determinar los haplogrupos de nueve (9) individuos (1.3%) debido a la baja calidad de las muestras de DNA. Aunque todas las poblaciones muestreadas tienen estructuras genéticas que se ajustan en líneas generales a los patrones que se podrían esperar para grupos indígenas contemporáneos de Centro y Suramérica, encontramos que los haplogrupos A y B eran más frecuentes en el norte de Colombia, mientras los haplogrupos C y D eran más frecuentes en el sur y suroccidente del país.
Palabras clave: mtDNA; nativos americanos; migraciones humanas; Colombia; Suramérica; haplogrupos, Método RFLP; Fragmentos de Restricción de Longitud Polimórfica
Resumo
A caracterização de DNA mitocondrial (mtDNA) permite o estabelecimento de estruturas genéticas e relações filogenéticas em populações humanas, rastreando linhagens ao longo do tempo. Nós analisamos amostras de mtDNA de vinte (20) populações Nativas Americanas (700 individuos) distribuidas ao longo do territòrio colombiano. Amostras foram coletadas durante 1989-1993 dentro do contexto do programa Expedido Humana ("Expedición Humana") e armazenados no Repositório Biológico do Instituto de Genética Humana (IGH) da Pontificia Universidad Javeriana (Bogotá, Colombia). Haplogrupos foram determinados por análise do polimorfismo no comprimento de fragmentos de restrição (RFLPs). O haplogrupo mais frequente foi o haplogrupo A, com 338 individuos (48,3%). Esse haplogrupo também é um dos mais frequentes em Mesoamérica, e interpretamos nossos resultados como um modelo de suporte que propóe que grupos que falam Chibchan migraram ao Norte de Colombia a partir da Mesoamérica ainda em tempos pré-históricos. Halogrupo C foi encontrado em 199 individuos (28,4%), enquanto que os menos frequentes foram B e D, com 113 e 41 (16% e 6%) individuos, respectivamente. Os haplogrupos de nove (9) individuos (1,3%) nào puderam ser determinados devido à baixa qualidade das amostras de DNA. Embora todas as populações amostradas tinham estruturas genéticas que cabem amplamente nos padróes esperados para os grupos indigenas contemporáneos da América Central e do Sul, foi observado que os haplogrupos A e B foram mais frequente no Norte da Colombia, enquanto que os haplogrupos C e D foram mais frequentes no Sul e Sudoeste da Colombia.
Palavras-chave: mtDNA; Nativos Americanos; migração humana; Colombia; América do Sul; haplogrupos, Método RLFP; Polimorfismo no Comprimento de Fragmentos de Restrição
Introduction
Traditionally, studies of the demographic history of human populations were based on archaeological records and historical documents. During the later 20th century, these approaches were supplemented by studies of classical genetic markers, such as ABO blood groups and HLA loci. More recently, it has become possible to use the direct analysis of autosomal DNA, the non-recombining portion of the Y chromosome (NRY DNA), and mitochondrial DNA (mtDNA) to study the genetic structure of human populations in greater detail. mtDNA, inherited through the maternal line, has certain properties that make it a valuable tool for analysing variability among human populations (Wallace et al. 1999, Ingman et al. 2000). Analysis of mtDNA haplogroups makes it possible to distinguish particular maternal lineages within populations, offering new opportunities for the comparison of these results with those from archaeological, anatomical, and linguistic approaches.
Peopling of the Americas
It has long been understood that the Americas were most likely populated by migration from Asia across what is now the Bering Strait. Genetic, anatomical, and archaeological studies have proposed various models regarding the origin and number of prehistoric migrations from Asia to the Americas (Stone & Stoneking 1998). Some have proposed a single wave of migration (Bonatto & Salzano 1997, Fagundes et al. 2008, Hooshiar Kashani et al. 2012), while others have supported two (Torroni et al. 1992, Horai et al. 1993, Pitblado 2011) or three (Greenberg et al. 1986, Reich et al. 2012) migratory waves. Moreover, previous work has determined the presence of the four principal founding mtDNA haplogroups (A, B, C, and D; X is also found in low frequencies) among contemporary Native Americans derived from Asian populations (Schurr et al. 1990, Torroni et al. 1992, 1993a, 1994, Horai et al. 1993, Brown et al. 1998, Malhi et al. 2003, Lewis et al. 2005). Analysis of mtDNA sequences has provided not only a high level of resolution, confirming the Asian origin of Native Americans, but also time estimates for migrations into the Americas from Asia (Luiselli et al. 2000, O'Rourke & Raff 2010, Hooshiar Kashani et al. 2012). It now seems probable that the first American populations left north-eastern Asia as much as 30,000 years before the present (ybp), perhaps pausing in Beringia for some 15,000 years, during which time new mutations separated the New World lineages from their Asian relations, before then moving into the Americas proper (Torroni et al. 1993a, 1993b, Forster et al. 1996, Tamm et al. 2007, Goebel et al. 2008, Hooshiar Kashani et al. 2012). It is possible that two further migrational pulses from Asia contributed to populations speaking Eskimo-Aleut languages and to the Canadian Chipewyan people, who speak a Na-Dene language (Goebel et al. 2008, Reich et al. 2012). The remainder of the Native American population, however, would seem to be descended from the initial Asian migration.
This scenario recalls Greenberg's controversial proposals (Greenberg et al. 1986, 1987, Greenberg & Ruhlen 1992), which elaborated on an theory originally developed by Sapir (1916) and which has since been sustained by Ruhlen (1994), that Native American languages can be divided in three linguistic macro-families: Amerind, Eskimo-Aleut, and Na Dene. This analysis has, however, been strongly criticized on methodological grounds (Bolnick et al. 2004), and the conservative consensus among contemporary linguists recognizes a large number of as-yet distinct American language families (Campbell 1997). South America alone is understood to contain some 108 separate language families and isolates (Campbell 2012, Muysken & O'Connor 2014). Although linguistic analysis can be a powerful tool to aid in reconstructing human prehistory (Amorim et al. 2013), and there has accordingly been much recent interest in comparing linguistic and genetics studies, it is critical to represent patterns of both genetic and linguistic variation accurately if such work is to contribute usefully to our understanding (Szathmary 1984). Moreover, it must be remembered that it is not uncommon for populations to shift languages—or for ethnicity (and language) to be shared across populations of diverse genetic heritage. In the case of American languages, analysis has long been hampered by poor and limited data, and reassessment of higher-level relationships may need to await the collection of more and better information. Nevertheless, in recent decades considerable progress has been made in clarifying the relationships between at least some American languages, offering fresh opportunities for comparison with the results of archaeological, anatomical, and genetic studies.
Peopling of South America
Regarding indigenous South American populations, studies of autosomal microsatellite markers have revealed a pattern of progressively reduced heterozygosity from north to south and from west to east (Wang et al. 2007). Most models for the peopling of South America take one of two basic forms. One of these assumes that an initial migration into the continent progressed south from Mesoamerica along the Andes, thereafter expanding eastward at various points. The other assumes that the initial migration from Mesoamerica split into easterly and westerly groups in the northern part of the continent, thereafter progressing southward separately (Bodner et al. 2012). There are also questions about the possibilities of later population pulses (Lewis et al. 2007), as in the case of Chibchan speakers (see below), who seem to have entered northern South America from Mesoamerica sometime during the Holocene (Constenla Umaña 1995, Melton et al. 2007). Ultimately, no real consensus on how South America was initially peopled has been reached. Research using data from modern populations is complicated by the fact that, although some populations (e.g. that of Venezuela) reveal relatively high frequencies of indigenous haplogroups, many indigenous South American populations have been impacted by significant inflows of European and African genes over the last 500 years (Salas et al. 2004, Mendizabal et al. 2008, Catelli et al. 2011, Gómez-Carballa et al. 2012).
Chibchan-speaking populations of Mesoamerica and northern South America
Modern speakers of Chibchan languages are dispersed across lower Mesoamerica and northern South America. In recent decades, a sound understanding of the Chibchan language family has been established (Constenla Umaña 1981, 1991, 2005, 2008, Quesada 2007) and complemented by archaeological studies (Lange & Stone 1984, Bray 2003, Hoopes & Fonseca Z 2003, Hoopes 2005) that, taken together, indicate Chibchan-speaking populations originated in Mesoamerica, spreading to northern South America in prehistoric times. Studies of mtDNA diversity amongst modern Mesoamerican Chibchan-speakers have found they are characterized by high frequencies of haplogroup A (>65%), markedly lower frequencies of B (20-30%), very low frequencies of D, and absence of C (Santos & Barrantes 1994, Torroni et al. 1994, Batista et al. 1995, Kolman et al. 1995, Kolman & Bermingham 1997). However, a study by Melton et al. (2007) shows that although modern Chibchan-speaking populations in Colombia's Sierra Nevada de Santa Marta (SNSM) likewise have high frequencies of haplogroup A (>65%), they also have moderate frequencies of C (<35%), and very low frequencies of B as well as D (<7.5%, if present). Thus, Chibchan populations seem most prominently characterized by haplogroup A (common in Mesoamerica) and moreover show strong genetic affinities with contemporary Mayan-speakers (Melton et al. 2007). The secondary frequency of haplogroup C distinguishes the Chibchan-speakers of Colombia's SNSM from Mesoamerican Chibchan-speakers, in whom B is found at similar frequencies. Haplogroup C is rare in other southern Mesoamerican populations (Kolman & Bermingham 1997) but common in other South American populations (Keyeux et al. 2002). Moreover, haplogroup D is common in northern South America (Lalueza-Fox 1996, Lalueza-Fox et al. 1997) though not amongst either Mesoamerican or Colombian SNSM Chibchan-speakers.
Indigenous populations of contemporary and prehistoric Colombia
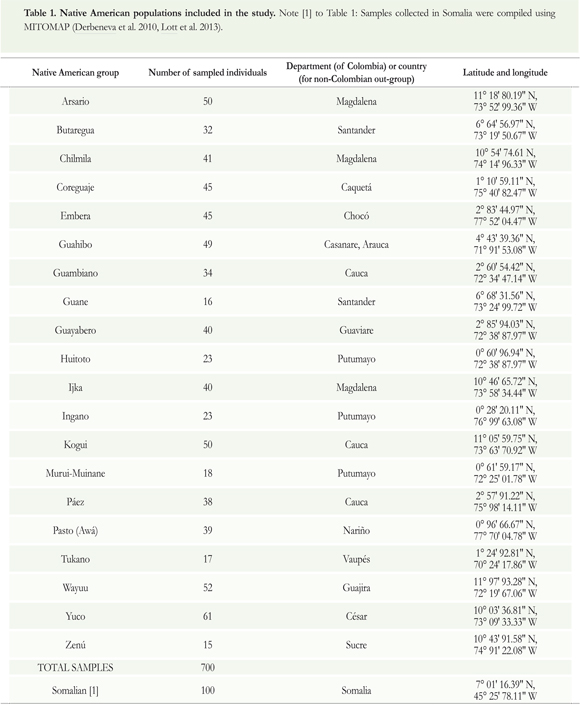
In Colombia, recent estimates suggest that around 68 indigenous American languages, classified into 13 language families and 8 language isolates, are still spoken by perhaps some 700,000 people (Landaburu 2005). Presently, the state recognizes over 80 ethnic minority groups (including Afro-Colombian and Romani-speaking groups), scattered throughout the country in both rural and urban contexts (Rojas et al. 2010). There is a considerable amount of variation in the scientific literature with regards to the nomenclature for American languages and ethnic groups. The present study uses (Spanish-language) names that align with the labelling practices of the Biological Repository of the Institute of Human Genetics (IGH) at the Pontificia Universidad Javeriana (Bogotá, Colombia), from which the samples used in the study were drawn. For guidance on correspondences between the names used in the present study and other common names frequently used to identify the same ethnic groups in English-language publications in linguistics and anthropology, see Supplementary material 3 (and, further, Adelaar & Muysken 2004 p 66).
Since the 1980s, genetic and clinical studies of molecular HLA antigens (Bernal et al. 1995), as well as of mtDNA and Y chromosomes (Keyeux et al. 2002, Melton et al. 2007, Rondón et al. 2008, Noguera 2012, Noguera et al. 2014), have shed fresh light on the genetic structures of Colombian populations, revealing a high degree of admixture between populations of different origins and a clear vision of antigenic diversity (Briceño et al. 1996). Some results from the analysis of nucleotide diversity amongst these groups suggest past population bottlenecks, followed by later recovery (Mitchell et al. 2000). Post-bottleneck populations tend to reveal low genetic variability, and the original proportion of alleles in the general population may have changed considerably. There can be significant frequencies of African haplogroups within modern Colombian indigenous populations, especially in areas bordering the Caribbean and Pacific coasts, and some effects of genetic isolation can be seen through high frequencies of mtDNA haplogroup L, attributed to the results of the trans-Atlantic slave trade (Nuñez et al. 2010). In contrast, northwestern Colombia reveals a high frequency of European Y chromosomes alongside a similarly high frequency of indigenous mtDNA (Carvajal-Carmona et al. 2000). Other studies suggest that some individuals lacking specific markers for major South American haplogroups could belong to haplogroup X, usually found in North American indigenous populations (Forster et al. 1996, Malhi & Smith 2002, Malhi et al. 2003, Ribeiro-dos-Santos et al. 2013).
Besides studies of modern populations, several studies of ancient Colombian DNA have recently been conducted. Remains from 11 individuals interred near Bogotá c. 2,000 ybp were found to belong exclusively to haplogroup B (Silva et al. 2008). In contrast, analysis of remains from 16 individuals from a nearby location in southwestern Bogotá, but dating from the immediate pre-Colonial period (c. 400-1000 ybp), revealed a mix of haplotypes A and B (Jara et al. 2010). Given the association of haplogroup A with both Central and South American Chibchan-speaking populations (Melton et al. 2007), this change in haplogroup patterns over time within the same geographical area has been associated with an influx of Chibchan speakers (Jara et al. 2010). In contrast, analysis of comparably aged (c. 860 ybp) remains from 17 individuals from the southern parts of the Department of Santander associated with the Colonial-era Guane people (of unknown linguistic affiliation) revealed higher frequencies of haplogroup B (41%), slightly lower frequencies of haplogroup A (35%), and lower frequencies of haplogroup D (24%) (Casas-Vargas et al. 2011). Accordingly, it may be that the (pre )Colonial Guane represented descendants of an earlier non-Chibchan-speaking population, characterized by high proportions of haplogroup B, that had more recently also received an influx of haplogroup A, perhaps from the neighboring Chibchan-speaking populations of the Altiplano Cundiboyacense. The Guane are known to have had trade relations with Chibchan-speaking Muisca of the (pre )Colonial Altiplano (Langebaek Rueda 1985, Lleras Pérez & Langebaek Rueda 1987, Pérez 1990) and conceivably had other kinds of relations with them as well.
The present study
Colombia occupies a pivotal juncture between Central and South America, where peoples from the Mesoamerican, Caribbean, Amazonian, and Andean regions have all interacted (Constenla Umaña 1991). Despite an increased pace of research in recent decades, many questions remain about the form and sequence of population movements, as well as prehistoric cultural and linguistic shifts, in the region. The present paper reports on a study that analyzed restriction fragment length polymorphisms (RFLPs) to identify the mtDNA haplogroups associated with various indigenous Colombian groups in order to establish their genetic structures and consider possible relationships between haplogroup frequencies and linguistic affiliations, particularly to reexamine proposed relationships between the extent of the Chibchan language family and high frequencies of haplogroup A. All results from this study were based only on the analysis of mtDNA haplogroups; haplotypes were determined only as a quality control.
Materials and methods
Population samples
For this study, we analyzed mtDNA from twenty (20) Native American populations dispersed throughout Colombia (Figure 1), sampling 700 individuals. The majority of the samples was collected during 19891993 within the context of the Expedición Humana project and stored in the Human Biological Repository of the Institute of Human Genetics (IHG), Pontificia Universidad Javeriana (Bogotá, Colombia). Another group of 282 samples had been collected separately from the Arsario, Coreguaje, Embera, Ijka, Kogui, and Wayuu populations by Ignacio Briceño in the context of his doctoral research (Briceño 1998), but these samples are included within the context of the Expedición Humana project (see Table 1 and Supplementary material 3). All human samples were obtained with informed consent and donated voluntarily. Data derived from samples collected in Somalia were compiled using MITOMAP (Derbeneva et al. 2010, Lott et al. 2013).

DNA extraction
DNA from the population samples was extracted through the phenol-chloroform method. Salting out was used for the extraction of the control samples, due to the ease of implementation, low toxicity of reagents, and facility of visualizing the DNA (Miller et al. 1988).
mtDNA analysis
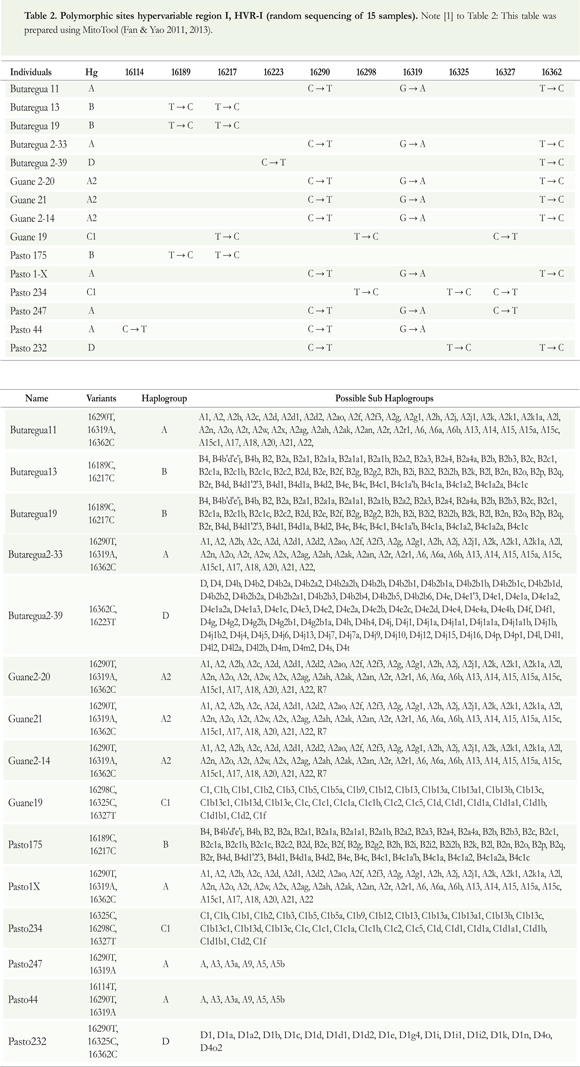
mtDNA was analyzed by RFLPs using primers based on Stone & Stoneking (1998) to identify haplogroups A, B, C, and D. In the present study, conditions for polymerase chain reaction (choice of template DNA, enzyme, dNTPs, magnesium chloride, and the necessary concentrations of each) were standardized. Hypervariable region I (HVR-I) was analyzed by sequencing DNA randomly for a small percentage (5%) as quality control. These samples were analyzed using primers based on Prieto et al. (2011) (see Table 2).
RFLP analysis defined existing haplogroups (the four founding haplogroups, A—D), according to three restriction sites, and a deletion in intergenic region 5. We analyzed the markers that define the founding Amerindian haplogroups as follows:
Haplogroup A. i) Restriction enzyme Hae III. ii) Site gain in nt663 and formation of fragments of 121, 66, 38, and 17 base pairs (bp).
Haplogroup B. i) 9-deletion base pairs [bp] in COII/tRNALys.
Haplogroup C. i) Restriction enzyme Hinc II. ii) DNA fragment of 181 base pair (bp), a loss of a HinclI site at position 13259, and formation of fragments of 155 and 26 bp.
Haplogroup D. i) Restriction enzyme Alu I. ii) DNA fragment of 183 base pair (bp) present cleavage site loss Alu site, position 5,176.
The amplification reaction to 700 samples had a final volume of 20 [μL, with 18.5 [μL of a master mix (Green Master Mix, Promega, 0.5 [iM of each primer (1[μL), and 1.5 [μL of sample. DNA amplification conditions proceeded in an initial denaturation at 94 °C for 15 minutes, followed by denaturation at 94 °C for 60 seconds, annealing at 51 °C for 60 seconds in haplogroup A, 54.3 °C for haplogroups B, C and D, and extension at 72 °C for 60 seconds. Final extension at 72 °C for 5 minutes was included. Each region was independently examined. PCR products were separated by electrophoresis in polyacrylamide gels and detected by ultraviolet irradiation after staining with ethidium bromide.
Data Analysis
F-statistics (Fst distances), were calculated using the Arlequin 3.5.1.2 software (Excoffier et al. 2005); see Supplementary material 1 and 2. An inter-analysis, based on the parameters of haplogroup diversity, was performed between each of the pairs of groups (individuals and/or populations) based on the methods of Tajima (1989) and Nei & Kumar (2000, ll. 284-286). We constructed a dendrogram outlining phylogeny or evolutionary kinship between the populations studied and in comparison to an African (Somali) population. This tree was constructed with the unweighted pair group method using arithmetic averages (UPGMA) (see, for example, Sneath & Sokal 1973), one of the most widely used methods of clustering of taxa or populations. Percentages of different mitochondrial haplogroups (see Figure 1) and Fst values for twenty (20) Colombian indigenous populations were calculated (see Table 3 and Supplementary material 3).

Alignment of sequences
Sequences were aligned with the revised Cambridge Reference Sequence (rCRS) (Anderson et al. 1981, Andrews et al. 1999) using MitoTool (Fan & Yao 2011, 2013). For each of the samples, changes were determined relative to the rCRS.
Results and discussion
Haplogroups, origins and affiliations of indigenous Colombia populations
Results of the analysis of mitochondrial DNA (see Table 2 and Figure 1) show that haplogroup A was most frequent, found in 338 individuals (48.3%), followed by haplogroup C, found in 199 individuals (28.4%). Less frequent were haplogroups B and D, found in 113 and 41 (16% and 6%) individuals, respectively. The haplogroups of nine (9) individuals (1.3%) could not be determined due to the low quality of the DNA. We found higher frequencies of haplogroups A and B in populations from northern (and mid-western) Colombia, and higher frequencies of haplogroups C and D in populations from southern Colombia (Figure 1). We constructed a phylogenetic tree to show the genetic distances between populations (Figure 2). The cladogram reveals four (4) genetically defined branches.
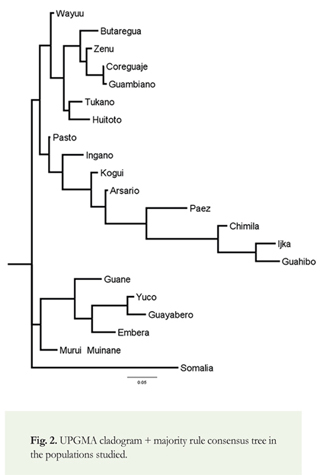
The first of these groups is characterized by the predominance of haplogroup C. It includes the Wayuu population (Arawakan-speaking), the Koreguaje and Tukano populations (Tucanoan-speaking), the Guambiano population (Barbacoan-speaking), the Huitoto population (speaking a Bora-Huitoto language), the Butaregua population (self-identified as descendants of the extinct Guane, whose language was of unknown affiliation), and the Zenú population (language extinct and of unknown affiliation). Here, it is significant to note the high frequency of C within the Arawakan-speaking Wayuu population, despite their close geographic proximity to the Chibchan-speaking populations of the SNSM (consisting of the Ijka, Kogui, and Arsario) in which haplogroup A predominates (Melton et al. 2007). The prevalence of C amongst the Wayuu may also support the notion that they share genetic affinity with other indigenous groups from the Amazonian region, where haplogroup C likewise predominates and in (or near) which the Arawakan languages are thought to have originated (Aikhenvald 1999, 2006; Walker & Ribeiro 2011).
The second group is characterized by the predominance of haplogroup A. It includes the Pasto (whose language is extinct, but thought to have probably belonged to the Barbacoan family) and Ingano populations (Quechuan-speaking), as well as the Ijka, Kogui, Arsario, and Chimila populations (all Chibchan-speaking), the Páez (Nasa Yuwe) population (speaking a language isolate), and the Guahibo population (speaking a Guahiban language). The Pasto population, located in southern Colombia, is characterized by higher frequencies of haplogroups A and C (38% and 28% respectively), as well as lower frequencies of haplogroups D and B (21% and 3%, respectively). The Pasto also appear to be genetically closest to the Huitoto population (of the first, C-dominated group), with a genetic distance of 0.006, and to the Ingano population, with a genetic distance of 0.007. The Pasto samples present a greater genetic distance from Ijka, Chimila, and Guahibo populations, despite likewise exhibiting a higher frequency of haplogroup A (possibly as a result of genetic influence from prehistoric Chibchan-speaking immigrants to the region). The Chibchan-speaking Chimila presented a high frequency of A (80%) with somewhat lower frequencies of B (20%) and no incidence of C or D, very similar to the results obtained by Batista et al. (1995) for the Chibchan-speaking Kuna [Cuna] of the Panamanian-Colombian border region (approximately 72% A, 28% B, with no C or D).
A third group is comprised of populations containing three or four founding haplogroups, and includes the Murui and Huitoto Muinane populations (speaking Bora-Huitoto languages), a modern self-identified Guane population, the Embera population (speaking a Chocoan language), the Guayabero population (Guajiboan-speaking) and the Yuco population (Cariban-speaking). Interestingly, the Guayabero population, living in the departments of Meta and Guaviare, speak a language of the same family as the neighboring Guahibo population, but their genetic profile, showing a high frequency of haplogroup B, is completely different from that of the Guahibo, who belong to the A-dominated second group identified in this study.
Finally, the dendrogram shows the distinct Somali African population. It was chosen for comparative purposes as it is genetically very distant from Colombian indigenous populations.
Additionally, we present the mutational changes (polymorphic sites of the mtDNA) of hypervariable region I at different positions in the sequence to determine founding haplogroups. These changes are compared (see Table 3) to the rCRS (Anderson et al. 1981, Andrews et al. 1999). We found that all individuals show the same haplogroups as determined by both methods. Using HVR-I allowed identification of haplotypes A2, B2, and C1 within a small subsample of individuals (5%) as a quality control; these are all specific haplotypes found in indigenous American populations but not in Asian or European populations.
The Chibchan-speaking populations of Colombia's SNSM (Ijka, Kogui, and Arsario) share similar profiles, with high frequencies of haplogroups A and C (and very low frequencies of B, if present, and no incidence of D). However, they are distinct from the Chibchan-speaking Kuna [Cuna] (Batista et al. 1995) and Chimila to their west, which in contrast both present modest frequencies of haplogroup B (28% and 20%, respectively) but no incidence of C (or D). The Wayuu, northeast of the SNSM, also present significant frequencies of A and C, though they were distinguished from the Chibchan-speaking SNSM populations by a somewhat higher frequency (23%) of haplogroup B (similar to the frequencies of that haplotype in the Chimila and Cuna [Kuna]). The nearby Cariban-speaking Yuco population, southeast of the SNSM near the Venezuelan border, has an even higher frequency (43%) of haplotype B. Archaeological and linguistic analysis suggests that Cariban languages may have spread both northwest and southeast from the Orinoco River delta in relatively late prehistory, perhaps from the thirteenth century AD (Muysken 2012). The Yuco language seems to represent part of a spread of Cariban inland from the Caribbean coast in prehistoric times (Adelaar & Muysken 2004), presumably at the expense of other languages, possibly including Chibchan. The Wayuu population also shows a genetic resemblance to Amazonian populations, many of which are likewise Arawakan-speaking. Archaeological and linguistic analyses suggest that the Arawakan languages, like the Cariban languages but in an earlier period, may have spread from a place of origin in the Amazon down the Orinoco valley to the Caribbean coast and into the Caribbean islands by 2,500 byp (Heckenberger 2013).
The close association between high frequencies of haplogroup A and Chibchan linguistic affiliation found in previous studies (Melton et al. 2007) suggests Colombian indigenous populations with elevated frequencies of haplogroup A may have been influenced by Chibchan-speaking populations or may have themselves shifted from Chibchan to other languages. Alternatively, as haplogroup A is moreover associated with Central American populations generally, the possibility that speakers of other unknown Mesoamerican language families migrated into Colombia in prehistoric times cannot be completely ruled out. In any event, the Guahibo present a high frequency of haplogroup A (86%), while in contrast their close geographical and linguistic neighbors the Guayabero have a markedly lower frequency of haplogroup A (48%). These data suggest a considerable genetic distance separates the two groups, and could support the hypothesis that the ancestors of the (now Guahiboan-speaking) Guahibo population had Mesoamerican and/or Chibchan-speaking origins or influences. Besides the strong genetic similarity of the Guahibo with Colombian Chibchan-speakers (particularly the Ijka of the Sierra Nevada de Santa Marta), it is noted that Guahibo inhabit a different habitat (savannah) than the Guayabero (jungle). We interpret the high frequency of haplogroup A in Guahibos as result of a founder effect among other possible mechanisms.
Samples from the Zenú population of the Department of Sucre, whose now extinct language is of unknown affiliation, revealed no presence of haplogroup B (comparable with the extremely low levels in the Chibchan-speakers of the Sierra Nevada de Santa Marta) and a lower (in comparison with those Chibchan-speakers) though still considerable proportion of haplogroup A (33%), but a relatively high proportion of haplogroup C (67%). This situation could result of a population bottleneck, as is the case in other groups revealing a high frequencies of haplogroup A, with low genetic diversity (i.e. Chibchan-speaking populations; Melton et al. 2007). It should be noted that the Zenú samples' pattern of high frequencies of C and somewhat lower frequencies of A sets them apart from their Chibchan-speaking neighbors in northern Colombia or in southern Panama (Batista et al. 1995).
The populations identified in this study as Butaregua (a settlement name) and Guane are both from near the town of Barichara in rural Santander. Moreover, both self-designate as indigenous descendants of the (pre) Colonial Guane population, though they are certainly admixed. Neither population has official state recognition as an ethnic minority. Samples from the modern Butaregua population reveal a particular combination of haplogroups A, B, and D (the most northerly extension of D found in our samples) reminiscent of (though not identical to) that found in ancient remains associated with the prehistoric Guane (A: 35%; B: 41%; D: 24%; Casas-Vargas et al. 2011). However, the Butaregua also reveal a significant frequency (52%) of haplogroup C, which is absent from the ancient Guane samples but otherwise common in varying proportions in many modern Colombian indigenous populations. The modern Guane population likewise contrasts with the ancient Guane in having a modest but marked frequency of haplogroup C. A further contrast between the modern Guane and both the ancient Guane and their modern Butaregua neighbors is the complete absence of haplogroup D. Clearly, the specific genetic histories of the modern Butaregua and Guane populations must differ significantly, despite both groups' claims of common descent from the ancient Guane. It is likely both modern populations have been impacted by genetic drift, founder effects, and/or population bottlenecks.
Amongst the Pasto, from the department of Nariño, haplogroup A was most frequent (38%), with somewhat lower frequencies of haplogroups C (28%) and D (21%). Although the Pasto language is extinct, it is thought very likely to have belonged to the Barbacoan family, which has more typological similarities with the central Andean sphere characterized by Quechuan than with the northern Colombian Chibchan sphere (Constenla Umaña 1991). Quechuan influence was strong in this region during immediate pre-Colonial times (after c. 1400 AD), but it is not clear whether the Andean affinities of Barbacoan hint at a deeper relationship with Quechuan or simply an earlier period of areal interaction (Adelaar & Muysken 2004). Nevertheless, it seems likely that the ancestors of the modern Pasto were poised, both linguistically and genetically, between the wider Andean/Quechuan and Chibchan spheres.
Conclusions
The results of the present study show that haplogroups A and B are more frequent in northern Colombia, while haplogroups C and D are more frequent in southern Colombia. This broadly supports the findings of previous studies (Lalueza-Fox 1996, Lalueza-Fox et al. 1997, Keyeux et al. 2002, Melton et al. 2007) that have found frequencies of haplogroup D approaching 100% in the Southern Cone of South America, while haplogroup A is very frequent in Central America (where D is relatively uncommon). Likewise, we conclude that the higher frequencies of haplogroup A in northern Colombia may be best explained as the result of prehistoric immigrations from Central America. Such migrations would have included at least a significant proportion of Chibchan speakers (as no other linguistic community of Central American origin has been shown to have expanded into northern South America). Although the genetic structures of most indigenous Colombian populations seem to fall broadly into the described northern A-B and southern C-D patterns, the Guahibo and Ijka populations present particularly high frequencies of haplogroup A (86% and 92%, respectively), suggesting that either genetic drift, founder effects and/or population bottlenecks have set them slightly apart.
Further research on mtDNA haplotypes from the same populations used in the present study should permit greater precision in determining the amount of mitochondrial genetic diversity within Colombian populations. It should be cautioned that further analyses of haplotypes within these populations may not necessarily support the present study's conclusions, which are based solely on haplogroups.
Nevertheless, the results of the present study contribute to the ongoing genetic analysis of modern Colombian indigenous populations, and offer significant new opportunities for comparison with ancient DNA from the region, though we would certainly recommend that further studies of ancient and modern population genetics be performed. Although comparative work across disciplines shows that populations defined through genetic analysis may sometimes correlate with populations identified by material culture and/or linguistic affiliation, though they also make it clear that various instances of prehistoric language shift also took place. Thus, simplistic equations of genetically defined populations with linguistically or culturally defined populations must be rejected unless stronger evidence for linking them can be presented. The analysis of such evidence would certainly call for greater interdisciplinary collaboration. Elsewhere, in both the Americas and Eurasia, models for interpreting persistentfrontiers (long-lasting conjunctions of varying combinations of various archaeologically, genetically, ecologically, and/or linguistically defined zones) have proven fruitful for interpreting prehistoric ethnolinguistic and/or cultural zones (Anthony 2007). Similar kinds of interdisciplinary approaches to collecting and analyzing new data should likewise be pursued in Colombia (and elsewhere in the Americas).
Acknowledgment
The Authors acknowledge to Institute of Human Genetics - Pontificia Universidad Javeriana for financing this study.
Conflict of interest
There are no conflicts of interest with funding sources or institutions.
References
Adelaar WFH, Muysken PC (2004) The languages of the Andes. Cambridge University Press, Cambridge. [ Links ]
Aikhenvald AY (1999) The Arawak language family. In: Dixon RMW Aikhenvald AY (eds) The Amazonian languages. Cambridge University Press, Cambridge, UK, pp 65-106. [ Links ]
Aikhenvald AY (2006) Arawak languages. In: Brown K (ed) Encyclopedia of language and linguistics. 2nd ed. Elsevier Science, New York, NY, pp 446-449 doi: 10.1016/B0-08-044854-2/02292-6. [ Links ]
Amorim CEG, Bisso-Machado R, Ramallo V, Bortolini MC, Bonatto SL et al. (2013) A Bayesian approach to genome/linguistic relationships in native South Americans. PloS one 8(5):e64099 doi: 10.1371/journal. pone.0064099. [ Links ]
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290(5806):457-465 doi: 10.1038/290457a0. [ Links ]
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM et al. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genetics 23(2):147 doi: 10.1038/13779. [ Links ]
Anthony DW (2007) The horse, the wheel and language: How Bronze-Age riders from the Eurasian steppe shaped the modern world. Princeton University Press, Princeton. [ Links ]
Batista O, Kolman CJ, Bermingham E (1995) Mitochondrial DNA diversity in the Kuna Amerinds of Panamá. Human Molecular Genetics 4(5):921-929 doi: 10.1093/hmg/4.5.921. [ Links ]
Bernal JE, Duran C, Briceño I, Ortega J, Papiha SS (1995) HLA antigens in five Amerindian groups (Yuko, Bari, Tunebo, Guane and Paez) of Colombia: Results of "Expedición Humana". Human Heredity 45(4):186-191 doi: 10.1159/000154287. [ Links ]
Bodner M, Perego UA, Huber G, Fendt L, Röck AW et al. (2012) Rapid coastal spread of first Americans: Novel insights from South America's Southern Cone mitochondrial genomes. Genome Research 22(5):811-20 doi: 10.1101/gr.131722.111. [ Links ]
Bolnick DA, Shook BA, Campbell L, Goddard I (2004) Problematic use of Greenberg's linguistic classification of the Americas in studies of Native American genetic variation. The American Journal of Human Genetics 75(3):519-522 doi: 10.1086/423452. [ Links ]
Bonatto SL, Salzano FM (1997) A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data. Proceedings of the National Academy of Sciences of the United States of America 94(5):1866-1871. [ Links ]
Bray W (2003) Gold, stone, and ideology: Symbols of power in the Tairona tradition of northern Colombia. In: Quilter J, Hoopes JW (eds) Gold and aower in Ancient Costa Rica, Panama, and Colombia: A symposium at Dumbarton Oaks, 9 and 10 October 1999. Dumbarton Oaks Research Library and Collection, Washington, DC, pp 301-344. [ Links ]
Briceño I (1998) Histocompatibility antigens and mitochondrial genome: serological and molecular genetic studies of Amerindians in Colombia. Doctorate thesis. Department of Biochemistry and Genetics, University of Newcastle upon Tyne, Newcastle upon Tyne, UK. [ Links ]
Briceño I, Gomez A, Bernal JE, Papiha SS (1996) HLA-DPB1 polymorphism in seven South American Indian tribes in Colombia. European Journal of Immunogenetics 23(3):235-40. [ Links ]
Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC et al. (1998) mtDNA haplogroup X: An ancient link between Europe/Western Asia and North America? American Journal of Human Genetics 63(6):1852-1861 doi: 10.1086/302155. [ Links ]
Campbell L (1997) American Indian languages: The historical linguistics of Native America. Oxford University Press, Oxford, UK. [ Links ]
Campbell L (2012) Classification of the indigenous languages of South America. In: Campbell L, Grondona V (eds) The indigenous languages of South America: A comprehensive guide. Walter de Gruyter, Berlin, Germany [ Links ]
Carvajal-Carmona LG, Soto ID, Pineda N, Ortíz-Barrientos D, Duque C et al. (2000) Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. American Journal of Human Genetics 67(5):1287-95 doi: 10.1016/S0002-9297(07)62956-5. [ Links ]
Casas-Vargas A, Gómez A, Briceño I, Díaz-Matallana M, Bernal JE et al. (2011) High genetic diversity on a sample of pre-Columbian bone remains from Guane territories in northwestern Colombia. American journal of physical anthropology 146(4):637-49 doi: 10.1002/ ajpa.21626. [ Links ]
Catelli M, Alvarez-Iglesias V, Gómez-Carballa A, Mosquera-Miguel A, Romanini C et al. (2011) The impact of modern migrations on present-day multi-ethnic Argentina as recorded on the mitochondrial DNA genome. BMC Genetics 12(1):77 doi: 10.1186/1471-2156-12-77. [ Links ]
Constenla Umaña A (1981) Comparative Chibchan phonology. Doctorate thesis. Department of Linguistics, University of Pennsylvania, Philadelphia, USA. [ Links ]
Constenla Umaña A (1991) Las lenguas del área intermedia: Introducción a su estudio areal. Editorial Universidad de Costa Rica, San José, Costa Rica. [ Links ]
Constenla Umaña A (1995) Sobre el estudio diacrónico de las lenguas chibchenses y conocimiento del pasado de sus hablantes. Boletín Museo del Oro 38-39:13-55. [ Links ]
Constenla Umaña A (2005) ¿Existe relación genealógica entre ls lenguas misumalpas y las chibchas? Estudios de Lingüística Chibcha 24:7-85. [ Links ]
Constenla UmañaA (2008) Estadoactual de lasubclasificación de las lenguas chibchenses y de la reconstrucción fonológica y gramatical del protochibchense. Estudios de Lingüística Chibcha 27:117-135. [ Links ]
Derbeneva O, Chalkia D, Lvova M, Hasseln K Von, Wallace DC (2010) 119 Mitochondrial DNA mutations found in native Central and South American samples provide evidence for mitochondrial adaptation to new environments. Mitochondrion 10(2):233 doi: 10.1016/j.mito.2009.12.110. [ Links ]
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47-50 doi: 10.1111/j.1755-0998.2010.02847.x. [ Links ]
Fagundes NJR, Kanitz R, Eckert R, Valls ACS, Bogo MR et al. (2008) Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. American Journal of Human Genetics 82(3):583-92 doi: 10.1016/j.ajhg.2007.11.013. [ Links ]
Fan L, Yao Y-G (2011) MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion 11(2):351-6 doi: 10.1016/j.mito.2010.09.013. [ Links ]
Fan L, Yao Y-G (2013) An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion 13(4):360-3 doi: 10.1016/j. mito.2013.04.011. [ Links ]
Forster P, Harding R, Torroni A, Bandelt HJ (1996) Origin and evolution of Native American mtDNA variation: A reappraisal. American Journal of Human Genetics 59(4):935-945. [ Links ]
Goebel T, Waters MR, O'Rourke DH (2008) The late Pleistocene dispersal of modern humans in the Americas. Science 319(5869):1497-502 doi: 10.1126/science.1153569. [ Links ]
Gómez-Carballa A, Ignacio-Veiga A, Alvarez-Iglesias V, Pastoriza-Mourelle A, Ruiz Y et al. (2012) A melting pot of multicontinental mtDNA lineages in admixed Venezuelans. American Journal of Physical Anthropology 147(1):78-87 doi: 10.1002/ajpa.21629. [ Links ]
Greenberg JH, Turner CGI, Zegura SL (1986) The settlement of the Americas: A comparison of the linguistic, dental, and genetic evidence. Current Anthropology 27(5) :477-497. [ Links ]
Greenberg JH (1987) Language in the Americas. Stanford University Press, Stanford, USA. [ Links ]
Greenberg JH, Ruhlen M (1992) Linguistic origins of Native Americans. Scientific American 267(5):94-99 doi: 10.1038/scienuficamerican1192-94. [ Links ]
Heckenberger Mi (2013) The Arawak diaspora: Perspectives from South America. In: Keegan WF, Hofman CL, Rodriguez Ramos R (eds) The Oxford handbook of Caribbean archaeology. Oxford University Press, Oxford, UK, pp 111-125. [ Links ]
Hoopes JW (2005) The emergence of social complexity in the Chibchan world of southern Central America and northern Colombia, AD 300-600. Journal of Archaeological Research 13(1):1-47 doi: 10.1007/s10814-005-0809-4. [ Links ]
Hoopes JW, Fonseca Z OM (2003) Goldwork and Chibchan identity: Endogenous change and diffuse unity in the Isthmo-Colombian area. In: Quilter J, Hoopes JW (eds) Gold and power in ancient Costa Rica, Panama, and Colombia: A symposium at Dumbarton Oaks, 9 and 10 October 1999. Dumbarton Oaks Research Library and Collection, Washington, DC, pp 49-89 [ Links ]
Hooshiar Kashani B, Perego UA, Olivieri A, Angerhofer N, Gandini F et al. (2012) Mitochondrial haplogroup C4c: a rare lineage entering America through the ice-free corridor? American Journal of Physical Anthropology 147(1):35-9 doi: 10.1002/ajpa.21614. [ Links ]
Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S et al. (1993) Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Molecular Biology and Evolution 10(1):23-47. [ Links ]
Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408(6813):708-13. doi: 10.1038/35047064. [ Links ]
Jara NP, Diaz M, Villegas V, López de Mesa C, Torres D et al. (2010) Aplication of authenticity criteria in mitochondrial studies on archaic bone remains from a prehispanic Muisca population. Colombia Médica 41(4):306-314. [ Links ]
Keyeux G, Rodas C, Gelvez N, Carter D (2002) Possible migration routes into South America deduced from mitochondrial DNA studies in Colombian Amerindian populations. Human Biology 74(2):211-33. [ Links ]
Kolman CJ, Bermingham E (1997) Mitochondrial and nuclear DNA diversity in the Chocó and Chibcha Amerinds of Panamá. Genetics 147(3):1289-302. [ Links ]
Kolman CJ, Bermingham E, Cooke R, Ward RH, Arias TD et al. (1995) Reduced mtDNA diversity in the Ngóbé Amerinds of Panamá. Genetics 140(1) :275-83. [ Links ]
Lalueza-Fox C (1996) Mitochondrial DNA haplogroups in four tribes from Tierra del Fuego - Patagonia: Inferences about the peopling of the Americas. Human Biology 68(6):853-871. [ Links ]
Lalueza-Fox C, Pérez-Pérez A, Prats E, Cornudella L, Turbón D (1997) Lack of founding Amerindian mitochondrial DNA lineages in extinct aborigines from Tierra del Fuego-Patagonia. Human Molecular Genetics 6(1):41-46 doi: 10.1093/hmg/6.1.41. [ Links ]
Landaburu J (2005) Las lenguas indígenas de Colombia: Presentación y estado del arte. Amerindia 29/30:3-22. [ Links ]
Lange F, Stone D (1984) The archeaology of lower Central America. University of New Mexico Press, Albuquerque, NM. [ Links ]
Langebaek Rueda CH (1985) Cuando los Muiscas diversificaron la agricultura y crearon el intercambio. Boletín Cultural y Bibliográfico 32(3):3-11. [ Links ]
Lewis CM, Lizárraga B, Tito RY, López PW, Iannacone GC et al. (2007) Mitochondrial DNA and the peopling of South America. Human Biology 79(2) :159-178 doi: 10.1353/hub.2007.0031. [ Links ]
Lewis CM, Tito RY, Lizárraga B, Stone AC (2005) Land, language, and loci: mtDNA in native Americans and the genetic history of Peru. American Journal of Physical Anthropology 127(3):351-360 doi: 10.1002/ajpa.20102. [ Links ]
Lleras Pérez R, Langebaek Rueda CH (1987) Producción agrícola y desarrollo sociopolítico entre los Chibchas de la Cordillera Oriental y Serranía de Mérida. In: Drennan R, C. Uribe (eds) Chiefdoms in the Americas. University Press of America, Lanham, MD, pp 251-270. [ Links ]
Lott MT, Leipzig JN, Derbeneva O, Michael Xie H, Chalkia D et al. (2013) MtDNA variation and analysis using MITOMAP and MITOMASTER. Current Protocols in Bioinformatics 1(123):1.23.1-26 doi: 10.1002/0471250953.bi0123s44. [ Links ]
Luiselli D, Simoni L, Tarazona-Santos E, Pastor S, Pettener D (2000) Genetic structure of Quechua-speakers of the Central Andes and geographic patterns of gene frequencies in South Amerindian populations. American Journal of Physical Anthropology 113(1) :5-17 doi: 10.1002/1096-8644(200009)113:1<5::AID-AJPA2>3.0.CO;2-U. [ Links ]
Malhi RS, Mortensen HM, Eshleman J a., Kemp BM, Lorenz JG et al. (2003) Native American mtDNA prehistory in the American Southwest. American Journal of Physical Anthropology 120(2):108-124 doi: 10.1002/ajpa.10138. [ Links ]
Malhi RS, Smith DG (2002) Brief communication: Haplogroup X confirmed in prehistoric North America. American Journal of Physical Anthropology 119(February):84-86 doi: 10.1002/ajpa.10106. [ Links ]
Melton PE, Briceño I, Gómez A, Devor EJ, Bernal JE et al. (2007) Biological relationship between Central and South American Chibchan speaking populations: Evidence from mtDNA. American Journal of Physical Anthropology 133(1):753-70 doi: 10.1002/ajpa.20581. [ Links ]
Mendizabal I, Sandoval K, Berniell-Lee G, Calafell F, Salas A et al. (2008) Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evolutionary Biology 8:213 doi: 10.1186/1471-2148-8-213. [ Links ]
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research 16(3):12-15 doi: 10.1093/ nar/16.3.1215. [ Links ]
Mitchell RJ, Federle L, Sofro AS, Papiha SS, Briceño I et al. (2000) Further data on the microsatellite locus D12S67 in worldwide populations: An unusual distribution of D12S67 alleles in Native Americans. Human Biology 72(4):697-705. [ Links ]
Muysken PC (2012) Contacts between indigenous languages in South America. In: Campbell L, Grondona V (eds) The indigenous languages of South America: A comprehensive guide. Walter de Gruyter, Berlin, Germany, pp 235-258. [ Links ]
Muysken PC, O'Connor L (2014) Introduction: South American indigenous languages - Genealogy, typology, contacts. In: O'Connor L, Muysken P (eds) The native languages of South America: Origins, development, typology. Cambridge University Press, Cambridge, UK, pp 1-28. [ Links ]
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford, UK. [ Links ]
Noguera MC (2012) Estudio molecular de estructura genética en poblaciones indígenas y afrocolombianas mediante marcadores nucleares y mitocondriales. Doctorate thesis. Faculty of Medicine, Pontificia Universidad Javeriana, Colombia. [ Links ]
Noguera MC, Schwegler A, Gomes V, Briceño I, Alvarez L et al. (2014) Colombia's racial crucible: Y chromosome evidence from six admixed communities in the Department of Bolivar. Annals of Human Biology 41(5):453-459 doi: 10.3109/03014460.2013.852244. [ Links ]
Nuñez C, Baeta M, Sosa C, Casalod Y, Ge J et al. (2010) Reconstructing the population history of Nicaragua by means of mtDNA, Y-chromosome STRs, and autosomal STR markers. American Journal of Physical Anthropology 143(4):591-600 doi: 10.1002/ajpa.21355. [ Links ]
O'Rourke DH, Raff JA (2010) The human genetic history of the Americas: The final frontier. Current Biology 20(4):R202-7 doi: 10.1016/j.cub.2009.11.051. [ Links ]
Pérez PF (1990) El comercio e intercambio de la coca: Una aproximación a la etnohistoria de Chicamocha. Boletín Museo del Oro 27:14-35. [ Links ]
Pitblado BL (2011) A tale of two migrations: Reconciling recent biological and archaeological evidence for the Pleistocene peopling of the Americas. Journal of Archaeological Research 19(4):327-375 doi: 10.1007/s10814-011-9049-y. [ Links ]
Prieto L, Zimmermann B, Goios a., Rodriguez-Monge a., Paneto GG et al. (2011) The GHEP-EMPOP collaboration on mtDNA population data - A new resource for forensic casework. Forensic Science International: Genetics 5(2):146-151 doi: 10.1016/j.fsigen.2010.10.013. [ Links ]
Quesada JD (2007) The Chibchan languages. Editorial Tecnológica de Costa Rica, Cartago, Costa Rica. [ Links ]
Reich D, Patterson N, Campbell D, Tandon A, Mazieres S et al. (2012) Reconstructing Native American population history. Nature 488(16):370-374 doi: 10.1038/nature11258. [ Links ]
Ribeiro-dos-Santos AM, De Souza JES, Almeida R, Alencar DO, Barbosa MS et al. (2013) High-throughput sequencing of a South American Amerindian. PLoS One 8(12):13-15 doi: 10.1371/journal.pone.0083340. [ Links ]
Rojas W, Parra MV, Campo O, Caro MA, Lopera JG et al. (2010) Genetic make up and structure of Colombian populations by means of uniparental and biparental DNA markers. American Journal of Physical Anthropology 143(1):13-20 doi: 10.1002/ajpa.21270. [ Links ]
Rondón F, Osorio JC, Peña ÁV, Garcés HA, Barreto G (2008) Diversidad genética en poblaciones humanas de dos regiones Colombianas. Colombia Médica 39(2):52-60. [ Links ]
Ruhlen M (1994) On the origin of languages: Studies in linguistic taxonomy. Stanford University Press, Stanford, CA. [ Links ]
Salas A, Richards M, Lareu M-V, Scozzari R, Coppa A et al. (2004) The African diaspora: Mitochondrial DNA and the Atlantic slave trade. American Journal of Human Genetics 74(3):454-465 doi: 10.1086/382194. [ Links ]
Santos M, Barrantes R (1994) D-loop mtDNA deletion as a unique marker of Chibchan Amerindians. American Journal of Human Genetics 55:413-414. [ Links ]
Sapir E (1916) Time perspective in aboriginal American culture: A study in method. Government Printing Bureau, Ottawa, Canada. [ Links ]
Schurr TG, Ballinger SW, Gan YY, Hodge JA, Merriwether DA et al. (1990) Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. American Journal of Human Genetics 46(3):613-623. [ Links ]
Silva A, Briceño I, Burgos J, Torres D, Villegas V et al. (2008) Análisis de ADN mitocondrial en una muestra de restos óseos arcaicos del periodo Herrera en la sabana de Bogotá. Biomédica 28(4):569-577. [ Links ]
Sneath PHA, Sokal RR (1973) Numerical taxonomy: The principles and practice of numerical classification. W H. Freeman & Company, San Francisco, CA. [ Links ]
Stone AC, Stoneking M (1998) mtDNA analysis of a prehistoric Oneota population: Implications for the peopling of the New World. American Journal of Human Genetics 62(5):1153-70 doi: 10.1086/301838. [ Links ]
Szathmary EJ (1984) Peopling of northern North America: Clues from genetic studies. Acta Anthropogenetica 8(1-2):79-109. [ Links ]
Tajima F (1989) The effect of change in population size on DNA polymorphism. Genetics 123(3):597-601. [ Links ]
Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG et al. (2007) Beringian standstill and spread of Native American founders. PloS One 2(9):e829 doi: 10.1371/journal.pone.0000829. [ Links ]
Torroni A, Schurr TG, Yang CC, Szathmary EJE, Williams RC et al. (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153-162. [ Links ]
Torroni A, Schurr TG, Cabell MF, Brown MD, Neel J V. et al. (1993a) Asian affinities and continental radiation of the four founding Native American mtDNAs. American Journal of Human Genetics 53(3):563-590. [ Links ]
Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF et al. (1993b) mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. American Journal of Human Genetics 53(3):591-608. [ Links ]
Torroni A, Neel J V, Barrantes R, Schurr TG, Wallace DC (1994) Mitochondrial DNA "clock" for the Amerinds and its implications for timing their entry into North America. Proceedings of the National Academy of Sciences of the United States of America 91(3):1158-1162 doi: 10.1073/pnas.91.3.1158. [ Links ]
Walker RS, Ribeiro LA (2011) Bayesian phylogeography of the Arawak expansion in lowland South America. Proceedings. Biological sciences / The Royal Society 278(1718):2562-2567 doi: 10.1098/rspb.2010.2579. [ Links ]
Wallace DC, Brown MD, Lott MT (1999) Mitochondrial DNA variation in human evolution and disease. Gene 238:211-30 doi: 10.1016/S0378-1119(99)00295-4. [ Links ]
Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N et al. (2007) Genetic variation and population structure in native Americans. PLoS Genetics 3(11):e185 doi: 10.1371/journal.pgen.0030185. [ Links ]













