Introduction
Healthcare-associated infections and antimicrobial resistance have increased in recent years to become a major worldwide healthcare issue, leading to high morbidity and mortality rates, as well as increased costs related to the treatment of infected patients1.
Healthcare-associated pathogens often contaminate both porous and nonporous inanimate surfaces of medical equipment, which act as potential reservoirs for these infectious agents. For this reason, there is a risk of indirect transmisión via these contaminated medical devices, which are reused on several patients and between procedures, or health professionals’ hands2,3.
Improving quality in healthcare involves enhancing the quality and safety of the medical devices used by health professionals in different complex procedures4. Contamination of medical devices has been identified in outbreaks and crosstransmission of pathogens among hospitalized patients in different clinical settings5-8. Contamination occurs either by transfer of microorganisms contaminating health workers’ hands or direct patient shedding of microorganisms into the equipment used during care delivery9.
Multidrug-resistant (MDR) bacteria have been reported as contaminating commonly used medical devices10. It has been reported that both Gram-positive and Gram-negative bacteria are able to survive up to months on dry inanimate surfaces, with longer persistence under humid and lowertemperature conditions10. Factors that may affect the transfer of microorganisms from one surface to another and crosscontamination rates are type of microorganisms, source and destination surfaces, humidity level, and size of inoculum11,12.
Therefore, medical device contamination is a major public health concern as the reusable medical devices are being extensively used for diagnostic and therapeutic purposes6. Several studies show that highly portable medical devices are associated with high contamination rates, often linked with bacterial cultures that are MDR to conventional antibiotic therapy7-9.
This challenge acquires a new dimension in the case of tourniquets used to perform invasive procedures involving peripheral venous puncture since these devices are applied proximal to the desired puncture site13-15. Consequently, tourniquets used in peripheral venous puncture can be a potential source of microbial contamination15. The noncompliance with the guidelines for managing medical devices can pose a risk for the dissemination of microorganisms16. For this purpose, it is recommended that tourniquets be manufactured using a material with a low risk for microbial contamination15,17. Most recent guidelines recommend the use of single-patient tourniquets17.
After an extensive review of the literature, no studies were found that synthesize information relative to the contamination of tourniquets used during procedures involving peripheral venous puncture.
Scoping review method
A scoping review was conducted based on the methodology proposed by the Joanna Briggs Institute for Scoping Reviews18,19. This review intends to answer the following question: What is the most common microbiological contamination found in tourniquets used by health professionals during peripheral venipuncture (contamination rate, microorganisms found and resistance profile)? The search strategy was limited to MEDLINE (via PubMed) and CINAHL complete (via EBSCO) databases. Studies written in English, Spanish, French, and Portuguese were considered for inclusion in this review, until the year 2017. The keywords as search query used in were ”Tourniquet” AND “Microbial contamination” OR “Bacterial colonization” OR “Microorganisms” OR “Infection” OR “Pathogens” OR “Fomites”. The study hits from the search strategy were reviewed for inclusion and exclusion criteria. Studies were considered eligible for inclusion if it was possible to evaluate the microbiological contamination of the tourniquets. Data extraction was guided by checklist assessing clarity of aims and research questions. The most relevant information was extracted from each eligible study (authors, year, country, sampled tourniquets, contaminated tourniquets, isolated bacteria and antibiotic resistance profile).
Results
A total of 1,587 studies were identified, of which 530 were duplicates. The remaining 1,057 studies were analyzed by title/abstract, and 36 were included for full-text analysis. Of these, only 20 were included in this review. Ten studies were excluded due to lack of microbiological data and six due to lack of access to the full-text version and author’s response (Figure 1). Of the studies included, eight were conducted in the United Kingdom20-27, two in Brazil28,29, and two in the United States of America30,31. Additionally, this review included studies conducted in Germany32, Australia33, South Korea34, Pakistan35, Portugal36, Nigeria37, New Zealand38 and Turkey39. The included studies were published between 1986 and 2017. A total of 1,477 reusable tourniquets were analyzed, ranging from 10 to 241 tourniquets analyzed per study and only 6 studies that had samples sizes ≥100. The microbial contamination rate varied of 9% to 100%. Relevant data such as the number of sampled tourniquets, the number of contaminated tourniquets, the main isolated bacteria per number of tourniquets and significant antibiotic resistance profiles are shown below (Table 1). The Staphylococcus genus was the most prevalent bacterial genus in the tourniquets contamination, the coagulase-negative staphylococci had high representativeness in all studies20-39. The Staphylococcus aureus was found in nineteen studies, demonstrated to be present, as contaminating agent, in more than 250 tourniquets, its prevalence, in these studies varied between 1-80%20-31,33-39. Eleven studies showed the presence of methicillin-resistant Staphylococcus aureus, varying the rate of contamination between 3.3-58.3%20-22,25,26,29,30,33,35,38,39.
Table 1 Characterization of the include studies relatively to the microbial contamination in tourniquets, isolated species and significant antibiotic susceptibility profile
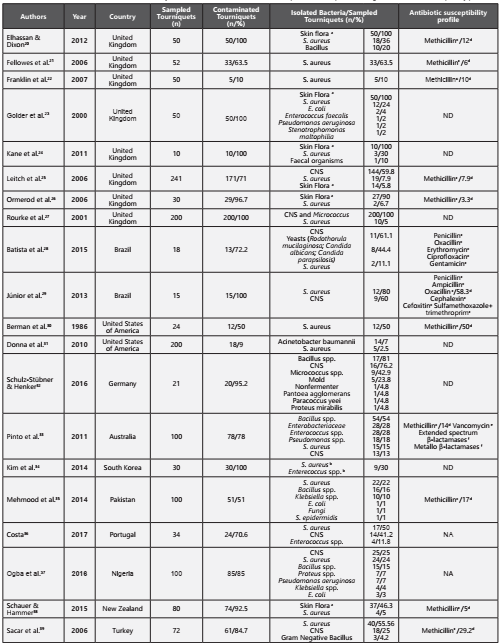
CNS - Coagulase-negative staphylococci; NA - Not available; ND - No detected methicillin-resistant strains aSkin Flora (several species in human skin); bSpecies quantified together per number of sampled tourniquets; cSusceptibility profile tested in Staphylococcus spp.; dPrevalence of Methicillin-resistant Staphylococcus aureus; e Susceptibility profile tested in Enterococcus spp.; f Susceptibility profile tested in Enterobacteriaceae.
In five studies, Enterococcus spp. was another microbial agent associated with tourniquets contamination, more tan 40 tourniquets23,24,33,34,36. Although only one study reported vancomycin-resistant Enterococci and it was found in 19 of contaminated tourniquets33.
In four studies, the genus Bacillus were found, responsible for 112 contaminated tourniquets20,32,33,35,37
Gram-negative bacteria were found in five studies, with 49 contaminated tourniquets23,32,33,35,37,39. Bacteria of the family Enterobacteriaceae namely Escherichia coli23,35,37, Klebsiella species35,37 and Proteus species32,37. Only one study reported contamination by Enterobacter cloacae resistant to extendedspectrum β-lactamases and metallo β-lactamases33.
Fungi were another important microbial group found in this review. In three studies28,32,35. Only one of the studies identified the type of fungi, confirming the presence of Rodothorula mucilaginosa, Candida albicans and Candida parapsilosis28.
Discussion
The main objective of this scoping review was to analyze and map studies examining the microbiological contamination of tourniquets used in peripheral venipuncture in order to understand the biological contamination in tourniquets and identify the most common microorganisms and their antibiotic susceptibility profile. Suspecting that the reusable tourniquets maybe important vehicles in the transmission of potential pathogens and since their site of use is close to the puncture site, many of the microorganisms present in there can be responsible for some bloodstream infections. Therefore we focused our microbiological analysis in the most relevant bacteria in this field.
Staphylococcus coagulase negative and S. aureus showed to be the most important pathogens contaminating tourniquets and other studies also emphasized its higher ability in survive for long periods of time in tourniquets, specially methicillin resistant species.We also observed a significant number of tourniquets contaminated with this type of the strains in several studies. Actually we know that the infections caused by this type of methicillin-resistant microorganisms are a serious public health issue due to the few therapeutic options available and their difficult eradication from hospital settings3,40-44. This resistance is mediated by gene mecA, which is carried by the mobile genetic element staphylococcal cassette chromosome mec (SCCmec). Methicillin-resistant Staphylococcus aureus produces an alternative penicillin-binding protein (PBP2A), which has low affinity for β-lactam antibiotics, resulting in oxacillin resistance. This resistance mechanism has been widely studied and is associated with several species of Staphylococcus both coagulase-negative and coagulase-positive40-44.
Other relevant species found to contaminate tourniquets was Enterococcus species. Besides of they are also able to survive in worst environmental conditions, this genus has increased considerably their antibiotic resistance making it difficult the treatment of enterococcal infections. The vancomycin-resistance is one of the most recently acquired resistances reported in this genus3,40,45,46. Glycopeptides, such as vancomycin, inhibit cell wall synthesis through their high affinity to the terminal d-alanyl-d-alanine group of peptidoglycan precursors. Until now, eight phenotypes of vancomycin-resistance have been identified, with the most common ones being vanA, vanB, and vanC. The manifestation of these phenotypes leads to changes in the peptidoglycan precursor, where the original D-alanyl-D-alanine terminus is replaced by Dalanyl-D-lactate or D-alanyl-D-serine, to which vancomycin has low binding affinity42,47-49. Only Pinto et al. in 2011 described the presence of vancomycin-resistant Enterococcus in tourniquets, however the other studies that detect this specie didn’t test the susceptibility profile of the strains23,24,33,34,36.
The Gram-negative bacteria namely Enterobacteriacceae was another group found in some studies as responsible to the contamination of tourniquets23,24,32,33,35,37,39. The production of β-lactamases is the most common mechanism of resistance studied in these family3,40,50-54. These enzymes are responsable for inactivating β-lactam antibiotics through the hydrolysis of their β-lactam ring. At present, the most worrying groups are the extended-spectrum β-lactamases and carbapenemases. The most common genes encoding extended-spectrum β-lactamases are blaTEM-1, blaSHV-1, and blaCTX-M. The most common in carbapenemases are encoded by the blaKPC, blaIMP, blaVIM, blaNDM, and blaOXA genes. These enzymes have an extended spectrum of activity and, when activated, complicate the treatment of the associated infections, reducing the therapeutic options available40,50-54. These types of antibiotic resistance mechanisms in isolates found contaminating tourniquets only were described in Pinto et al., in 2011 specifically in Enterobacter cloaceae. However, E. coli e Klebsiella normally carried out these genes and they are found in three studies23,35,37.
Fungal healthcare-associated infections have become a major challenge in clinical settings worldwide, with high morbidity and mortality rates55-57. The genus Candida is the leading cause of fungal infections in hospital environments, with values close to 80%55,56. Although Candida albicans is the most frequently isolated species in patients with invasive fungal infections, over the last decade, the epidemiology of this fungal genus has been changing, with an increase in the prevalence of the non-albicans species. This fact may be associated with the overuse of antibiotics and antifungals and increase of antibiotic resistance caused by bacterial adaptation, which complicate the treatment of this type of infections55-57. Three included studies in these review reported contamination of tourniquets, with fungi and yeast, specifically in Batista et al. 2015 that identified C. albicans and C. parapsilosis28,32,35.
Thus, these results show that reusable tourniquets used during clinical procedures are reservoirs of pathogens and they are moved from arm to arm and should be changed or disinfected regularly in order to ensure the safety and quality of healthcare. They showed to be potential fomites, proving with intervention studies, their possible contribution to healthcare-associated infections, specifically the bloodstream infections, disposable tourniquets may be more recommended.
Conclusion
This review intended to map studies that focus on the microbiological contamination of tourniquets used during peripheral venous puncture.
In the included studies, the contamination rate ranged from 9% to 100%, with 15 studies reporting rates equal or higher than 70%. The microorganisms responsible for these high contamination rates belong to different species, with the most prevalent being the Staphylococcus genus, followed by the Bacillus, Enterobacteriaceae family and Enterococcus species.
Nineteen studies identified Staphylococcus aureus as a common contaminant of tourniquets with more than 250 contaminated. The presence of resistance to methicillin was often associated with this species, in eleven studies the prevalence varied between 3.3-58.3%.
Patient safety can be at risk due to the high contamination rates found in tourniquets and their use in invasive procedures, such as peripheral venipuncture. For this reason, we suggest the disinfection procedure regularly in these medical devices or to adopt the disposable tourniquets. However, it will important provide evidences that the strains found in tourniquets are the same present in the catheter tips involved in bloodstream infections of the patient.















