INTRODUCTION
Colombia has an approximate cultivated area of 87,638 ha of citrus, encompassing orange, mandarin, and acid lime crops (MADR, 2021). However, its production faces various technological limitations, with one of the most significant challenges being the scarce availability of high-quality planting material, free from graft-transmitted diseases.
Various pathogens causing systemic diseases threaten citrus production, including Citrus tristeza virus (CTV) (Closterovirus: Closteroviridae) (Dawson et al., 2015), Candidatus Liberibacter asiaticus (Candidatus Liberibacter: Rhizobiaceae) (Bové, 2006) Citrus exocortis viroid (CEVd) (Pospiviroid: Pospiviroidae) (Semancik and Weathers, 1972), Citrus dwarfing viroid (CDVd) (Apscaviroid: Pospiviroidae) (Rakowski et al., 1994) and Hop stunt viroid (HSVd) (Hostuviroid: Pospiviroidae) (Ohno et al., 1983). All these pathogens have been reported in Colombia (Murcia et al., 2010; Murcia et al., 2020).
Among viroid-caused diseases, cachexia, formerly known as xyloporosis, stands out, with its causal agent being HSVd (Serra et al., 2018). Viroids are infectious agents consisting of circular RNA, single-stranded molecules without coat protein, of variable length ranging from 250 to 400 nucleotides (Venkataraman et al., 2021). Viroid replication depends on factors and enzymes encoded by the host since they lack the cellular machinery necessary for this process (Serra et al., 2018). Biologically, HSVd has been classified into pathogenic variants, types CVd-IIb and CVd-IIc (Serra et al., 2008), capable of inducing cachexia symptoms, and non-pathogenic variants, type CVd-IIa, which infect citrus plants without inducing symptoms (Semancik et al., 1988). It has been established that the type and severity of citrus cachexia symptoms depend on co-infections with other citrus viroids in the tree (Vernière et al., 2004). In susceptible hosts, a negative impact on yield has been observed, with reductions ranging from 34 to 76% and a decrease in fruit diameter (Belabess et al., 2021). The primary mode of spread is through grafting with buds infected with the viroid and the use of non-disinfected pruning tools. No vectors are known to be involved in the transmission of this disease (Duran-Vila, 2004; Barbosa et al., 2005).
Cases of cachexia affecting various citrus species have been reported worldwide, including in Iran (Teymuri et al., 2022; Zeitooni et al., 2023), China (Wang et al., 2010), the United States (Reanwarakorn and Semancik, 1999), and Brazil (Eiras et al., 2013). In Colombia, HSVd infection has been detected in Tahiti acid lime (Citrus x latifolia Tan.) in open field nurseries and commercial orchards (Murcia et al., 2010; Mosquera et al., 2015; Rodríguez-Mora et al., 2015). However, all identified isolates lacked the cachexia expression motif, indicating that they were non-pathogenic variants (Murcia et al., 2010; Mosquera et al., 2015).
Citrus cultivars affected by cachexia may exhibit visible symptoms or be asymptomatic (Vamenani et al., 2014; Belabess et al., 2021). In susceptible hosts such as Tangelo (C. reticulata Bla. × C. paradisi Macf.), symptoms include wood cracking, corresponding to wounds on the cambial face of the cortex, accompanied by discoloration of the phloem and gummy exudations (Duran-Vila et al., 2000). In advanced stages of the disease, plants may develop stem pitting on the trunk, as well as leaf chlorosis and stunning (Serra et al., 2008). Under favorable conditions for symptom manifestation, this disease can have fatal consequences for plants (Reanwarakorn and Semancik, 1988). During the nursery stage, infected plants are asymptomatic, making it necessary to guaranteehealthiness of planting material.
The detection of pathogens causing systemic diseases, including viroids, is carried out through biological methods and molecular techniques. For the biological diagnosis of citrus viroid diseases, the Arizona 861-S1 clonal Etrog citron (C. medica L.) is employed as an indicator plant, as recommended by Camps et al. (2014). This method involves observing the indicator plant's responses to viroid infection. For molecular diagnosis, reverse transcription polymerase chain reaction (RT-PCR) is used, as indicated by Guerrero et al. (2013). RT-PCR enables specific amplification of the viroid genome, providing a precise and efficient tool to confirm the presence of the infectious agent. The combination of biological and molecular methods offers a comprehensive approach to the diagnosis of systemic diseases, facilitating more precise and rapid viroid detection.
Managing viroid-induced diseases, such as citrus cachexia, requires the implementation of preventive measures, including the utilization of disease-free propagation material produced within a certification program that mandates compliance with molecular and biological diagnostics (NAPPO, 2013). Since 2019, Colombia has regulated the production of citrus plants in protected environments, classifying the multiplication of propagative material and seeds into four categories: 1) genetic, 2) foundation, 3) registered, and 4) certified (ICA, 2019). The foundation category represents propagative material obtained from genetic seeds, serving as the origin for the registered and certified seed categories. The Corporación Colombiana de Investigación Agropecuaria - Agrosavia, Centro de Investigación Palmira, maintains a collection of citrus varieties obtained through the micrografting method. These varieties are preserved under aphid-proof mesh greenhouse conditions and belong to the foundation seed category. Plants in this category are used as mother plants in the country's citrus certification program. The aim of this research was to assess the healthiness of citrus propagative material in the foundation category, comprising 17 citrus cultivars, against HSVd. This evaluation was conducted through both biological and molecular diagnosis, so that it can be used as a reliable source of buds intended for plant multiplication in the registered and certified seed category, contributing to maintaining quality and healthiness in citrus nurseries in Colombia.
MATERIALS AND METHODS
Plant material
The study was conducted in December 2021 at Corporación Colombiana de Investigación Agropecuaria - Agrosavia, Centro de Investigación Palmira, in the department of Valle del Cauca, Colombia. Within the center's facilities, 17 citrus varieties planted in pots have been maintained under controlled conditions inside an aphid-proof mesh greenhouse. These varieties are part of the elite citrus collection, obtained through in vitro micrografting of shoot apices. The plant collection consists of tangelos, grapefruits or pomelos, mandarins, oranges, and acid limes (Tab. 1). Each variety is represented by four replicates, totaling 68 plants.
Table 1. Collection of 17 micrografted citrus varieties.
| No | Common name | Scientific name |
|---|---|---|
| 1 | Minneola tangelo | C. reticulata Bla. × C. paradisi Macf. |
| 2 | Orlando tangelo | C. reticulata Bla. × C. paradisi Macf. |
| 3 | Clemenules mandarin | C. clementina |
| 4 | ICA Bolo mandarin | C. reticulata Bla. |
| 5 | Oneco mandarin | C. reticulata Bla. |
| 6 | Arrayana mandarin | C. reticulata Bla. |
| 7 | Campbell Valencia orange | C. sinensis (L.) Osb. |
| 8 | Frost Valencia orange | C. sinensis (L.) Osb. |
| 9 | García Valencia orange | C. sinensis (L.) Osb. |
| 10 | Olinda Valencia orange | C. sinensis (L.) Osb. |
| 11 | Salustiana orange | C. sinensis (L.) Osb. |
| 12 | Sweety orange | C. sinensis (L.) Osb. |
| 13 | Frost Washington orange | C. sinensis (L.) Osb. |
| 14 | Valle Washington orange | C. sinensis (L.) Osb. |
| 15 | Key lime | C. × aurantiifolia (Christm.) Swing. |
| 16 | Tahiti lime | C. × latifolia Tan. |
| 17 | Pomelo | C. paradisi Macf. |
Biological diagnosis
For biological indexing, the Etrog citron (C. medica L.) clone Arizona 861-S1, grafted onto the rootstock Volkameriana (C. volkameriana Ten. and Pasq.), served as an indicator plant. When Etrog citron plants reached 25 cm (from the graft union point), they were inoculated through vascular tissue contact with cortex from the 68 plants comprising the elite citrus collection, corresponding to the 17 micrografted citrus varieties. Branches from each quadrant were taken for each citrus variety, and these branches were used to inoculate the Etrog citron plants with two cortex segments each (Fig. 1). Four replicates of each citrus variety were inoculated into three Etrog citron plants. The experimental setup included negative controls, which consisted of three healthy non-inoculated Etrog citron plants obtained by micrografting of shoot apices, and positive control, which consisted of three healthy Etrog citron plants inoculated with an HSVd isolate obtained from the viroid strain collection of Agrosavia, conserved in vivo and previously confirmed by RT-PCR and sequencing (Mosquera et al., 2015).
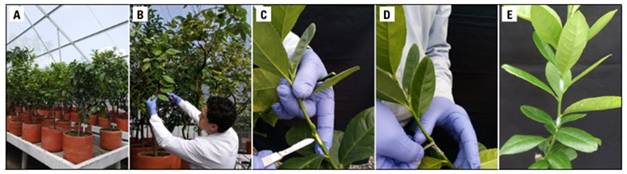
Figure 1. Biological indexing of 17 citrus varieties on Etrog citron (C. medica L.). A) collection of micrografted citrus varieties, preserved in an aphid-proof mesh greenhouse at Agrosavia, Centro de Investigación Palmira. B) collection of young shoots from citrus varieties. C) removal of the Etrog citron cortex for inoculation, and D) inoculation of Etrog citron with citrus cortex segments. E) Etrog citron plant inoculated with two cortex segments. Photo: Research Group - Agrosavia, Centro de Investigación Palmira.
The inoculated plants were transferred to an automated mesh greenhouse where suitable environmental conditions were maintained (temperature between 28 and 32°C) to promote viroid multiplication (Duran-Vila, 1989). Visual inspections were conducted every 15 d over 6 months to monitor the expression of disease symptoms.
Molecular diagnosis
Six months after inoculation (MAI), plant tissue was collected from Etrog citron plants inoculated with cortex segments from the 17 micrografted citrus varieties, as well as tissue from the negative control (non-inoculated Etrog citron plant) and from the positive control infected with HSVd. Each sample consisted of a pool of three plants, with four leaves per plant taken, totaling 70 samples (68 samples corresponding to the 17 citrus varieties, one negative control, and one positive control). The samples were packed in properly labeled paper bags and transported to the molecular genetic laboratory for processing.
RNA extraction
RNA extraction was performed using 100 mg of the leaf midrib tissue pulverized with liquid nitrogen, following the CTAB protocol described by Chang et al. (1993). To this tissue, 900 µL of CTAB extraction buffer was added [2% CTAB (Cetyl Trimethyl Ammonium Bromide), 2% PVP (polyvinylpyrrolidone), 100 mM Tris-HCl (pH 8), 25 mM EDTA, 2 M NaCl, 0.5 g L-1 spermidine, 2% β-mercaptoethanol], and the mixture was placed in a water bath at 65°C for 15 min. The samples were shaken every 5 min. Next, 1 mL of chloroform: isoamyl alcohol (24:1) was added, vortexed for 10 s, and centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was transferred to a new tube, and the process was repeated. The aqueous phase was transferred to a new tube, and 1 mL of 4 M LiCl was added. The samples were incubated overnight at 4°C. They were then centrifuged at 12,000 rpm for 30 min at 4°C, the supernatant was discarded, and the pellet was resuspended in 500 µL of TE-SDS buffer at 37°C. Subsequently, 700 µL of isopropanol and 200 µL of 5M NaCl were added, the tube was inverted several times, and it was incubated at -20°C for 1 h. The sample was centrifuged at 12,000 rpm for 10 min at 4°C, the supernatant was discarded, and the pellet was resuspended in 500 µL of 70% ethanol. After centrifugation at 12,000 rpm for 15 min at 4°C, the supernatant was discarded, and the pellet was dried for 1 h at room temperature in a laminar flow cabinet. The pellet was resuspended in 30 µL of RNase-free water and stored at -80°C. The quality and quantity of RNA in the samples were assessed using spectrophotometry with the NanoDropTM 1000 (ThermoFisher), analyzing the 260/280 and 260/230 absorbance ratios.
RT-PCR
Complementary DNA (cDNA) synthesis was carried out following the instructions of the SuperScript® III First-Strand Synthesis System for RT-PCR kit, catalog No. 18080-051 (Invitrogen). A mixture of 210 ng of RNA, 5.5 ng of random primer, and 0.83 mM of dNTPs (total) was denatured at 95°C for 5 min, resulting in a final volume of 9 μL. The reaction was then placed on ice for 1 min, and 5 µL of a mixture containing 2X RT Buffer, 10 mM MgCl2, 20 mM DTT, 20 U RNaseOUT, and 100 U SuperScriptTM III RT was added. Next, the reaction was incubated at 60ºC for 1 h. The reaction was terminated at 85ºC for 5 min, placed on ice for 1 min, and 1 U of Escherichia coli RNase H was added, followed by incubation at 37°C for 20 min. The resulting cDNA was stored at -20°C.
The quality of the cDNA was verified by amplifying the 18S ribosomal RNA (rRNA) region using the primers 18SF (5' GAGAAACGGCTACCACATCCA 3') and 18SR (5' CGTGCCATCCCAAAGTCCAAC 3'), following the conditions described by Du et al. (2006) with some modifications. The reaction setup consisted of 1X Taq buffer (KCl), 1.5 mM of MgCl2, 0.2 mM of dNTPs mixture, 0.2 µM of each primer, 0.5 U of Taq DNA polymerase (ThermoScientific) in a final volume of 12.5 µL. For the detection of HSVd, the specific primer combination HSVd/F2 (5'- GTGGCATCACCTCTCGGTT -3') and HSVd/R1 (5'-GGGGCTCCTTTCTCAGGTAAGTC -3') was used, following the conditions specified by Bernad and Duran-Vila (2006) with some modification. The reaction was setup with 1X Taq buffer (KCl), 1.5 mM of MgCl2, 0.5 mM of dNTPs mixture, 0.2 µM of each primer, 0.5 U of Taq DNA polymerase (ThermoScientific) in a final volume of 12.5 µL.
The amplification process was carried out in an Agilent Technologies SureCycler 8800 with the following programs: for 18S amplification an initial denaturation of 95°C for 5 min was followed by 35 cycles of 95°C for 30 s, 52°C for 60 s and 72°C for 60 s, with a final elongation step of 72°C for 10 min and a cool-down to 20°C for 5 min. For HSVd/F2-R1, an initial denaturation at 94°C for 3 min was followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 45 s, with a final elongation step of 72°C for 10 min and a cool-down to 20°C for 5 min. The amplified products were visualized on a 1.6% (w/v) agarose gel, run in 0.5X TBE buffer at 100 V, 400 mA for 80 min. Nucleic acids were stained with GelRed® (Biotium) at a concentration of 1.3X added to the PCR products, and visualization was done using the Enduro GDS gel documentation system (Labnet).
RESULTS AND DISCUSSION
Biological diagnosis
The indicator plants of Etrog citron clone Arizona 861-S1, inoculated with cortex tissue segments from the 17 micrografted citrus varieties, were compared with reference controls: the negative control (non-inoculated Etrog citron plant) and the positive control infected with the HSVd viroid. This method allowed the expression of symptoms in the host when the viroid infection occurred.
At 2 MAI, initial symptoms of HSVd infection were observed in the three inoculated replicates of positive control, characterized by petiole necrosis. At 4 MAI, petiole necrosis and leaf epinasty were evident. At 5 MAI, symptoms progressed to include petiole necrosis, stem necrosis, leaf epinasty, and basal and middle necrosis of the central leaf vein. Finally, at 6 MAI, a more advanced stage was reached, with leaf epinasty (Fig. 2A), petiole necrosis (Fig. 2B), stem necrosis (Fig. 2C), and necrosis of the central leaf vein at the basal, middle, and apical parts observed (Fig. 2D). These symptoms have been associated with infections caused by viroids in Etrog citron (Duran-Vila et al., 1988; Camps et al., 2014; Zeitooni et al., 2023). The plants used as negative controls (non-infected plants) (Fig. 2E, F, G, H) and Etrog citron plants inoculated with tissue segments from the 17 citrus varieties corresponding to the foundation seed (Fig. 2I, J, K, L) did not show any symptoms during the evaluation period. The presence of symptoms in positive controls, along with their absence in negative controls, demonstrates the quality of the biological indexing setup.
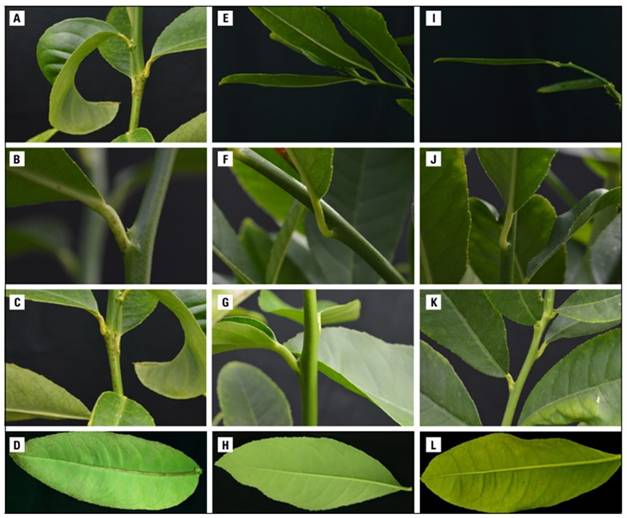
Figure 2. Biological indexing of HSVd in Etrog citron (C. medica) clone Arizona 861 S1, 6 months after inoculation. Positive control: A, B, C and D. Negative control: E, F, G and H. Inoculation of tissue from Minneola tangelo: I, J, K and L. Photo: Research group-Agrosavia, Centro de Investigación Palmira.
The Etrog citron clone Arizona 861-S1 is considered one of the best hosts for conducting biological diagnosis of viroids due to its high susceptibility and symptom expression for most viroids reported in citrus (Duran-Vila et al., 1988; Villalobos et al., 2004; Camps et al., 2014; Murcia et al., 2020; Rodríguez-Mora et al., 2022). This method achieves viroid multiplication at detectable titers by molecular techniques, confirming the results and providing greater specificity in diagnosis (Duran-Vila et al., 1993). Therefore, the results obtained in this evaluation were confirmed using molecular methods, specifically RT-PCR, as a complementary diagnostic approach.
Molecular diagnosis
The RNA extraction performed using the protocol developed by Chang et al. (1993) yielded concentrations ranging from 564.7 to 2,698.42 ng µL-1. The 260/280 absorbance ratios ranged from 1.99 to 2.23, and the 260/230 absorbance ratios varied between 1.92 and 2.41 (Fig. 3). These values are considered within the optimal ranges for use in laboratory practices, according to guidelines established by Koetsier and Cantor (2019).
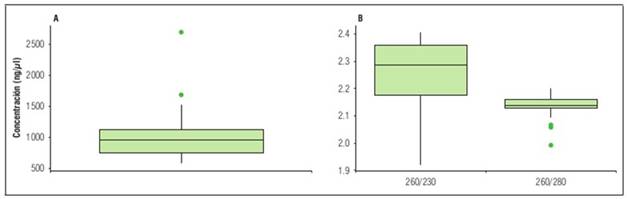
Figure 3 Box plots of quantification and quality of RNA extracted from Etrog citron using the protocol of 6, measured by spectrophotometry. A) concentration. B) purity ratios A260/280 and A260/230.
Amplification of the 18S ribosomal RNA (rRNA) region produced a fragment of approximately 250 base pairs (bp) in all analyzed samples (Fig. 4A). This result is consistent with the size reported by Du et al. (2006), indicating a synthesis of high-quality cDNA. In the positive control (healthy Etrog citron plants inoculated with an HSVd isolate), RT-PCR allowed the amplification of a region of the HSVd viroid of approximately 174 bp, which comprises the entire TL and P domains and partially the C domain, as reported by Bernad and Duran-Vila (2006). Non-specific amplified products exceeding the expected size were not included in the diagnosis. The viroid was not detected in either the 17 citrus varieties evaluated or the negative control (non-inoculated Etrog citron plant) (Fig. 4B). Thereby, the results obtained in the molecular diagnosis confirmed the healthiness in the 17 citrus varieties, consistent with the results obtained in the biological diagnosis.
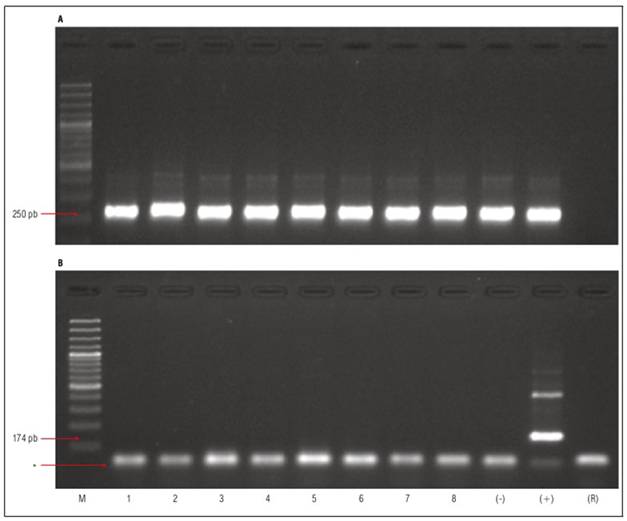
Figure 4. Detection of 18S rRNA and HSVd by RT-PCR. A) Amplification of the 18S rRNA region. B) HSVd amplification by RT-PCR. M: Molecular weight marker 100 bp (ThermoScientifc ref. #SM0323). (-): Negative control. (+): Positive control. (R): PCR reaction control without template. Lane 1: Minneola Tangelo, Line 2: Orlando Tangelo, Line 3: Clemenules mandarin, Line 4: ICA Bolo mandarin, Line 5: Sweety Orange, Line 6: Key lime, Line 7: Tahiti lime, Line 8: Pomelo. * Primer dimer.
RT-PCR is the most widely used method for diagnosing viroids in citrus, whether performed singly or in multiplex (Wang et al., 2009; Camps et al., 2014; Lin et al., 2015; Rodríguez-Mora et al., 2015). This technique is fast, economical, and effective, contributing to the improved detection and characterization of viroids affecting citrus. It also allows for the discrimination between pathogenic and non-pathogenic variants of HSVd (Bernad and Duran-Vila, 2006). The use of RT-PCR extends to indexing procedures, serving as an essential methodology for the early detection of viroids in citrus within certification programs (NAPPO, 2013).
Detecting HSVd infection in citrus cultivars poses significant challenges since infected plants can be asymptomatic and go unnoticed. Therefore, the application of both biological and molecular diagnostic methods becomes crucial for reliably monitoring the healthiness of citrus planting material.
This study confirmed the absence of HSVd in the citrus propagation material within the foundationcategory, including tangelos, grapefruits, mandarins, oranges, and acid limes. These varieties were maintained under controlled conditions within an anti-aphid mesh greenhouse at the Corporación Colombiana de Investigación Agropecuaria - Agrosavia, Centro de Investigación Palmira. Previous studies by Rodríguez-Mora et al. (2017) reported that this collection is free from systemic diseases such as tristeza, exocortis, and citrus huanglongbing, all of which are regulated in the Colombian citrus certification standard (ICA, 2019). This achievement represents a significant milestone and provides substantial support to Colombia's citrus certification program.
CONCLUSION
This study confirmed the healthiness of the citrus variety collection within the foundation category regarding HSVd. This implies the potential use of this material as a reliable source of buds for plant multiplication within the registered and certified seed category. Thus, it actively contributes to maintaining quality and health standards in citrus nurseries in Colombia. This represents a significant step towards ensuring the healthy and sustainable production of new citrus cultivated areas.















