Introduction
The neotropical legume genus Leucaena comprises 24 species, with a native range spanning the southern USA to northern Peru. Leucaena are mostly small trees (occasionally shrubs) with bipinnately compound leaves, a lack of stem or leaf armament, extrafloral nectaries, globose to subglobose inflorescences of many small flowers and elongate, flattened, dehiscent pods. Numerous human uses for Leucaena have contributed to a long history of use in Mesoamerica for food, shade, firewood and even spiritual medicine. Archaeological evidence from seed remains in caves dates the use of Leucaena seeds as a minor food source by Mixtec and Nahuatl people to at least 6,000 years ago (Zárate 2000) and seeds of 13 species have been recorded to be consumed in modern times in south-central Mexico (Hughes et al. 2007), where they are referred to as ‘guaje’ [goo-ah’-hay].
Research on Leucaena has focused on the archaeological history of plant use, patterns of evolutionary diversification among species, impacts of human use on diversification, as well as many applied research questions associated with multipurpose use in subsistence farming, modern agriculture and range management systems. The number of distinct species within the Leucaena genus has been investigated through reciprocal illumination of morphological and molecular evidence that currently supports the recognition of 19 diploid and 5 allotetraploid species (Hughes 1998a; Govindarajulu et al. 2011a). Studies into the underlying patterns of divergence and reticulation among species have involved phylogenetic, geographic and crossability projects. The morphological and ecophysiological diversity within the genus combined with high crossability among species provide ample opportunities for genetic improvement via traditional breeding approaches, and notably via interspecific artificial hybridization to develop genetically improved seed lines (Brewbaker et al. 1989; Brewbaker and Sorensson 1990). Additionally, newly generated genomic data, tools and resources are helping to advance our understanding of species relationships and will be critical for underpinning future basic and applied research on this interesting and economically important genus.
Here we provide an overview of species diversity and the evolutionary history of Leucaena, focusing especially on studies over the last 30 years. These studies have revealed a complex history, which includes paleopolyploidy, genomic diploidization, allopatric diploid divergences, recent interspecific hybridization and allopolyploidization precipitated by anthropogenic translocation and cultivation (Hughes et al. 2007; Govindarajulu et al. 2011a, 2011b). This complex myriad of evolutionary mechanisms influencing the history of Leucaena presents challenges for reconstructing an accurate phylogeny. In addition to reviewing past work on Leucaena, we summarize on-going and recently published genomic work and the utility of these new genomic data for basic and applied research on Leucaena.
Taxonomy and morphology
The genus Leucaena was first described in 1842 when Bentham (1842) transferred Acacia glauca, A. pulverulenta, A. diversifolia and A. trichodes to this newly recognized genus. Subsequent work by Bentham (1846; 1875), Standley (1922), Britton and Rose (1928), Brewbaker (1987a), Harris et al. (1994), Zárate (1994), Hughes (1998a) and Govindarajulu et al. (2011a) has all contributed to the modern circumscription of 24 species within the Leucaena genus.
The genus is placed in the informal Leucaena group alongside Desmanthus, Kanaloa and Schleinitzia within the mimosoid clade of the newly re-circumscribed legume subfamily Caesalpinioideae (LPWG 2017). Anther and pollen morphology as well as chloroplast and nrDNA ITS sequence data have been used to determine generic relationships within the Leucaena group (Hughes 1997; 1998b), which is part of a larger clade including the informal Dichrostachys group, plus the genera Prosopidastrum, Piptadeniopsis and Mimozyganthus (Hughes et al. 2003; Luckow et al. 2005; LPWG 2017).
All 24 species of Leucaena are woody, single or multi-stemmed trees or shrubs ranging from 4 to 25 m tall. The shoots are always free of spines or prickles. Terminal shoots can either be terete or ridged with corky fiber bundles. Leaves in Leucaena are always stipulate, alternate and bipinnate, but show significant and conspicuous quantitative variation within and between species in terms of numbers of pairs of pinnae per leaf and leaflets per pinna and leaflet size. Many species exhibit nyctinasty (circadian-based ‘sleep’ movement) in their leaflets and pinnae; however, seismonasty (touch sensitivity) does not occur in Leucaena (Hughes 1998a).
Extrafloral nectaries are found on various parts of the leaves of all Leucaena species. These nectar-secreting glands mediate mutualisms with ants for protection against herbivory, and are common across the majority of mimosoid legume genera (Marazzi et al. 2013). The morphology and arrangement of these structures help distinguish some Leucaena species from others (Hughes 1998a).
The stamen filaments are generally yellow, white or pink, and flowers are borne in globose or subglobose capitula (head-like clusters) that are variously arranged on flowering shoots. Pods generally arise in clusters of 1‒15, but sometimes as many as 45 from a single capitulum. Leucaena seeds typically have circular to ovate or ellipsoid shape and are dorsi-ventrally flattened (Hughes 1998a).
Cladistic analyses of morphological data (Hughes 1998a) revealed limited support for a number of groups. For example, the Leucaena esculenta group shares thick and corky bark with gray-metallic surfacing, while the closely related L. retusa and L. greggii share stipitate extrafloral nectaries. Quantitative analyses of leaf traits (number of pairs of pinnae, number of pairs of leaflets and size of leaflets) show clear patterns of morphological intermediacy in hybrids, including a dosage effect due to ploidy (Sorensson 1993; Hughes and Harris 1994, 1998; Hughes 1998b), suggesting tight genetic control of quantitative leaf morphology.
Crossability among species
Through a massive series of artificial intra- and interspecific crossing experiments, Sorensson and Brewbaker (1994) investigated the potential to generate hybrids as well as the mechanisms and degree of incompatibility within and among species. At that time, just 16 species (15 published and 1 unpublished) were recognized in the genus. Of the 120 possible 2-way mating combinations, 118 were artificially hybridized and 31 of the 32 possible self- and interspecific mating combinations tested (Sorensson and Brewbaker 1994). An impressive 58,218 floret emasculations and hand-pollinations were made, with 77% of 118 two-way combinations and 61% of 232 one-way combinations producing viable seed, demonstrating high crossability among species and the tremendous scope for the use of crossing in breeding work to generate novel hybrids, which have dominated Leucaena improvement programs to date (Brewbaker et al. 1989; Brewbaker and Sorensson 1990). Furthermore, the predominant factor in interspecific incompatibility was variation in ploidy between parents, whereas gametophytic self-incompatibility was noted at the intraspecific diploid level. Crossability among morphologically, genetically, geographically and even chromosomally distinct diploid species is consistent with a predominant mode of allopatric, rather than sympatric, speciation (Govindarajulu et al. 2011a).
Variation in chromosome number and genome size: Paleopolyploidy, diploidization and neopolyploidy
Most diploid mimosoids have a chromosome complement of 2n = 26 (e.g. Santos et al. 2012), suggesting a base number of x = 13 for mimosoids. However, the ‘diploid’ species of Leucaena, whose chromosome numbers have been counted, have 2n = 52 or 56 (Pan and Brewbaker 1988; Palomino et al. 1995; Cardoso et al. 2000; Schifino-Wittmann et al. 2000), which is consistent with Leucaena having experienced an ancient polyploidization (paleopolyploidization), i.e. whole genome duplication, prior to the diversification of the modern ‘diploid’ lineages. Nevertheless, these species are typically referred to as ‘diploids’ because they show primarily disomic, rather than tetrasomic, patterns of inheritance (Pan 1985; Sorensson and Brewbaker 1989).
Furthermore, genome size data (Palomino et al. 1995; Hartman et al. 2000; Govindarajulu et al. 2011b) for 24 species of Leucaena suggest that L. macrophylla has the smallest genome of all legumes (data.kew.org/cvalues) and that other diploid Leucaena species also have relatively small genomes ranging from 0.31 to 1.65 pg/1C. Although some of the absolute sizes of these genomes are inconsistent with subsequent unpublished estimates for all 19 diploid taxa (Trujillo and Bailey, unpublished data), both sources are consistent with typical ‘diploid’ genomes rather than full tetraploid complements that might have been retained from the paleopolyploidization event. Ultimately the combined evidence from chromosome numbers, disomic inheritance and genome sizes suggests extensive genomic diploidization following an ancestral paleopolyploidization along the stem lineage of Leucaena (Govindarajulu et al. 2011b; Figure 1).
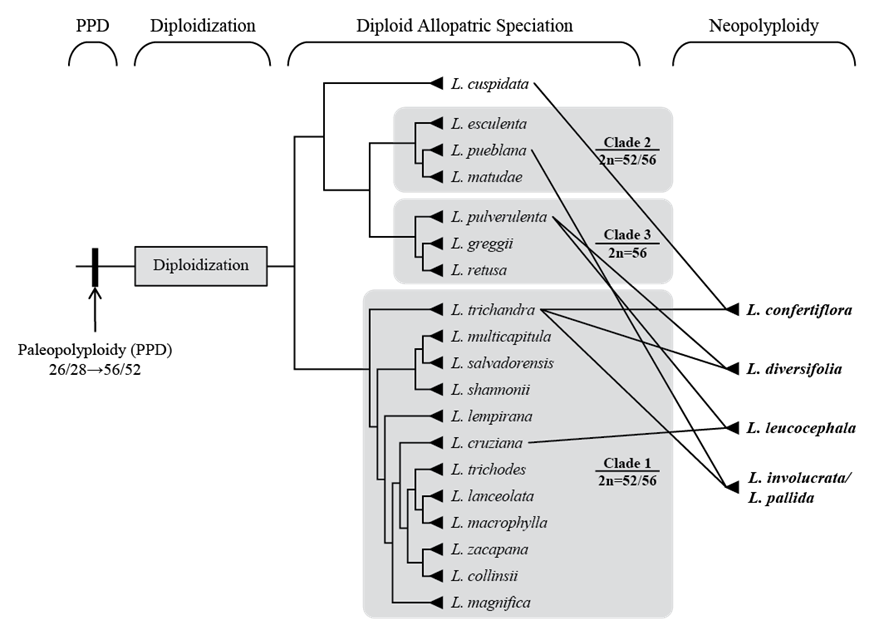
Figure 1 The evolutionary dynamics of polyploidy in Leucaena. The diploid phylogeny summarizes relationships recovered by Govindarajulu et al. (2011a). Multiple accessions of each species are collapsed to single terminals. Figure reprinted from Govindarajulu et al. (2011b) with permission from Wiley Company.
By contrast, the tetraploid Leucaena species have 2n = 104 or 112 chromosomes with genome sizes (Palomino et al. 1995; Hartman et al. 2000; Govindarajulu et al. 2011b) close to the sum of their parental complements (see below), consistent with little diploidization in the modern ‘tetraploid’ lineages, suggesting that either the mechanism of diploidization is not functioning to any great degree in these tetraploid lines, or these tetraploid taxa arose recently, offering insufficient time for diploidization to have significantly reduced genome sizes (Govindarajulu et al. 2011b).
Phylogenetics of Leucaena
Early assessments of relationships among species of Leucaena involved analysis of morphological, cytological and crossability evidence (Zárate 1984, 1994; Brewbaker 1987b; Pan and Brewbaker 1988; Hughes 1998b). The first molecular phylogenetic investigation of Leucaena (Harris et al. 1994) used cpDNA RFLP data for 22 species and showed for the first time 3 main clades of diploids. However, conflict between the cpDNA gene tree and morphology and cytology suggested that cpDNA might have been influenced by plastome capture, raising doubts about this initial cpDNA gene tree as a species tree.
A clearer understanding of species limits plus the addition of nrDNA ITS sequence data and a rescoring of the cpDNA restriction fragment length polymorphism (RFLP) data (Hughes et al. 2002) presented relationships that were in agreement with the previous cpDNA RFLP study (Harris et al. 1994) in resolving 3 main clades of diploid species. However, within these 3 clades, bootstrap support values (particularly in Clade 1) remained low. To address this problem, an approach based on random amplification of polymorphic DNA was used to develop a set of anonymous low-copy nuclear loci and these were sequenced (Bailey et al. 2004) to further estimate relationships among species (Hughes et al. 2007; Govindarajulu et al. 2011a, 2011b). Govindarajulu et al. (2011a), using 59 accessions representing all diploid taxa, recovered the 3 clades established from earlier molecular work as well as a more robust estimate of interspecific relationships. Results from this analysis also provided strong evidence for allopatric divergence as the predominant mode of speciation among the diploid species (as noted above).
In these studies, multiple diploid populations were sampled using AFLPs to explore species boundaries on a scale not possible with morphological or cytological characters alone (Govindarajulu et al. 2011a). The resulting population genetic results supported the previously recognized taxonomy (Hughes 1998a), except for L. lanceolata, which was shown to be polyphyletic leading to the addition of L. cruziana as a species distinct from L. lanceolata, and upranking of L. collinsii subsp. zacapana as a distinct species (L. zacapana) (Govindarajulu et al. 2011a).
With these clarifications of species limits and the accumulated phylogenetic evidence, there is strong support for recognizing 3 major clades of diploids: Clade 1 (L. collinsii, L. cruziana, L. lanceolata, L. lempirana, L. macrophylla, L. magnifica, L. multicapitula, L. salvadorensis, L. shannonii, L. trichandra, L. trichodes and L. zacapana); Clade 2 (L. esculenta, L. matudae and L. pueblana); and Clade 3 (L. greggii, L. retusa and L. pulverulenta), with little evidence of homoploid hybridization among or within these clades (Govindarajulu et al. 2011a). The position of the other diploid species L. cuspidata and the relationships among closely related species in Clade 1 remain poorly resolved, but forthcoming phylogenetic analyses using much larger DNA sequence data sets (plastomes and nuclear genes from transcriptomes) across species will likely resolve these last remaining phylogenetic questions. With a few minor exceptions, the 3 diploid clades occupy largely allopatric distributions: Clade 1, the most widespread, is distributed from northern South America through Central America, south-central Mexico and along the Pacific coast of Mexico as far north as Sonora in lowland seasonally dry tropical forests; Clade 2 is found in inland regions of the south-central Mexican highlands and seasonally dry valleys mainly south of the Mexican volcanic axis; and Clade 3 has the most northerly distribution in northeast Mexico (north of the central volcanic axis) extending into southern Texas and adjacent New Mexico in the USA.
Serendipitous hybridization and polyploidy
Allopatric distributions of diploid sister species are consistent with geographical isolation and predominantly allopatric diploid speciation (Govindarajulu et al. 2011a). However, all 5 tetraploid species of Leucaena show clear evidence of hybrid (i.e. allopolyploid) origins, implying sympatry of their putative diploid parental species, but sympatry appears to be rare among wild diploid populations. Indeed, evidence suggests that each allotetraploid resulted from crosses between species placed in different diploid clades, which themselves have distinct geographies, further emphasizing the lack of sympatry among diploid species in the wild (Govindarajulu et al. 2011b).
Figure 2 illustrates the hypothesized origins of each allotetraploid species (Govindarajulu et al. 2011b). Here we refer to parental lines in terms of extant species; however, if the crosses were much older, the parental line would have been akin to, but not necessarily the same as, the modern species. Three of the 5 allotetraploids include L. trichandra as the putative paternal diploid parent crossed with L. pulverulenta to form tetraploid L. diversifolia and a species of the L. esculenta group to form L. involucrata and L. pallida. While L. pallida and L. involucrata may have the same polyploid origin, minor differences in morphology as well as allopatric distributions suggest these are 2 distinct species. The fourth L. trichandra-derived tetraploid, L. confertiflora, has L. trichandra as the putative maternal parent and L. cuspidata as the paternal line. Repeated involvement of L. trichandra in the origins of 4 of the 5 tetraploids, particularly on the paternal side, is consistent with its high propensity to produce unreduced pollen grains, its wide geographical distribution and early signs of its use as a human food source (Govindarajulu et al. 2011b). The fifth tetraploid species, the widely translocated and pantropically cultivated and naturalized L. leucocephala is derived maternally and paternally from L. pulverulenta and L. cruziana, respectively.
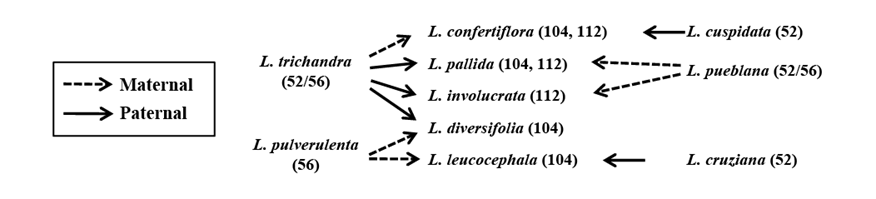
Figure 2 Inferred pattern of tetraploid origins from diploid progenitors. Maternal origins are based on findings from cpDNA and congruent resolution in nuclear-derived gene trees. Paternal origins derive from the divergent placement of nuclear-derived sequences in comparison with the inferred maternal origins. Figures reprinted from Govindarajulu et al. (2011b) with permission from Wiley Company.
The divergent hybrid origin of each tetraploid lineage has raised interesting questions about the likely origin(s) of these taxa. The available evidence suggests that at least some of these tetraploid species may be the product of serendipitous backyard hybridization via juxtaposition in informal cultivation in central Mexico over the last 6,000 years (Hughes et al. 2007; Govindarajulu et al. 2011b). Evidence consistent with this anthropogenic backyard allopolyploid formation hypothesis includes the aforementioned predominance of allopatry among wild diploids, limited genomic diploidization of tetraploids suggesting recency of the tetraploids (see above), and archeological evidence that suggests the oldest seeds of tetraploid Leucaena date from about 1,500 years ago, long after the first appearance of Leucaena seed remains, this despite the predominant cultivation and use of allotetraploids in backyard gardens today (Hughes et al. 2007; Govindarajulu et al. 2011b).
Several other putative spontaneous polyploid hybrids have been discovered and documented across south-central Mexico. These are also thought to have arisen following juxtaposition of their parents in cultivation. Two of these have been named as hybrid species. First, the named hybrid taxon, L. ×mixtec, a putative triploid between tetraploid L. leucocephala and diploid L. esculenta, is a relatively common tree across south-central Mexico (Hughes and Harris 1998). As expected for a triploid, these L. ×mixtec hybrids are sterile and each individual thus likely represents a de novo F1 spontaneous hybrid. A second named hybrid taxon, L. ×spontanea, a putative hybrid between tetraploid L. leucocepahala and L. diversifolia, occurs as scattered individuals wherever these 2 species occur together (Hughes and Harris 1998). Finally, a few individuals of a putative hybrid between L. leucocephala and L. confertiflora have been documented, also in south-central Mexico (Hughes et al. 2007). The full extent of spontaneous interspecific hybridization in south-central Mexico remains to be fully investigated.
As a wider range of Leucaena species are cultivated on an ever wider scale, continued spontaneous hybridization and generation of new hybrids are likely, adding further complexity to an already complex picture of polyploidy and interspecific hybridization.
Developing genomic resources
Future applied and basic research on Leucaena will benefit greatly from recent and ongoing research to sequence a Leucaena genome and generate transcriptome data for all species. Diploid L. trichandra was selected as the species for genome sequencing because it is the putative progenitor of 4 of the 5 tetraploid species. Table 1 summarizes available Leucaena-associated NCBI Sequence Read Archive (SRA) resources, including genomic DNA reads representing organellar and nuclear genomes, as well as a variety of transcriptomic data (RNA-seq) from multiple species. Below we briefly review some of the pertinent findings from these studies and outline ongoing work.
The chloroplast genomes for Leucaena trichandra and other mimosoids were sequenced by Schwarz et al. (2015) and Dugas et al. (2015), resulting in resources relevant to the understanding of variation in coding and non-coding chloroplast sequence, cpDNA genome structure and RNA editing. Similarly, Kovar et al. (2018) recently published a mitochondrial genome (L. trichandra) with discussion on the origin of mtDNA DNA variation and mitochondrial RNA-editing across the genus. In these studies, the mimosoid organellar genome(s) is considerably larger than their papilionoid legume counterparts. The authors discuss some of the sources and mechanisms behind this size variation. The associated RNA-seq and gDNA-seq genomes are essential prerequisites for the development of organellar-derived species-specific markers and for gene expression studies at the organellar level.
In addition to organellar genomic resources, the Bailey Lab at New Mexico State University is currently completing a draft nuclear genome, based on PacBio and Illumina sequence data for the diploid L. trichandra (Bailey et al. unpublished data - available, with stipulations on publication priority/conflicts from the authors on request). Current analysis and annotations on the genome suggest that L. trichandra, and presumably other ‘diploid’ Leucaena, retain considerable evidence of the paleotetraploidization event that predates the divergence of ‘diploid’ Leucaena (Figure 1).
In addition to these resources, the Bailey Lab (NMSU) and the Borthakur Lab (UNH Manoa) are continuing to work on a number of resources, including an investigation of plant transcript response to psyllid feeding in L. cruziana (Lakshman et al. in prep.) and the Bailey Lab has transcriptomic and raw genomic data available from Leucaena psyllids. Like some of their relatives (www.ncbi.nlm.nih.gov/genome/genomes/867?genome_assembly_id=31561), these psyllid genomes display considerable bias in GC content that complicates the use of Illumina data to assemble a full genome.
Future directions
These new genomic data are providing new insights into the phylogenetic relationships among diploid species (Abair et al. in preparation) plus the origins of the allotetraploids, as well as tools and resources for germplasm improvement. While the basic phylogenetic framework and likely polyploid parentages are now fairly well established, the details of these polyploid origins in terms of where, when and how many times they happened remain poorly understood. The emerging genomic data resources provide access to unlimited genetic markers that could be used to test for multiple independent origins for each of the 5 Leucaena tetraploids, and most notably the globally important L. leucocephala and its morphologically variable taxonomic subspecies. A key element in future work is likely to involve much denser sampling of accessions of tetraploids and their diploid parents to fully reveal the complexities of this extensive hybrid and polyploidy series.
These new genomic tools and resources, alongside a better understanding of the evolutionary history of Leucaena, also present exciting new opportunities for Leucaena genetic improvement and breeding programs, including efforts to develop seed or sterile lines with low potential for invasiveness, decreased mimosine concentration and traits that improve their utility in difficult environments (salinity, cold, drought, etc.).
















