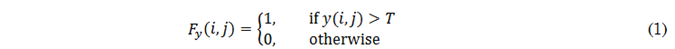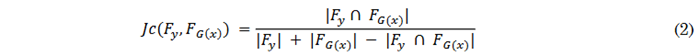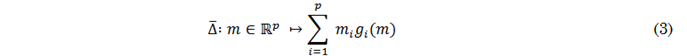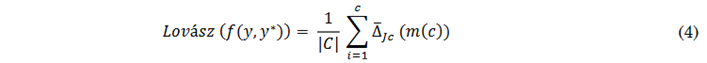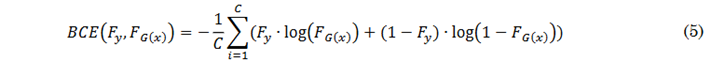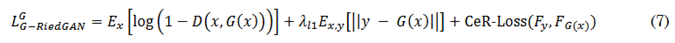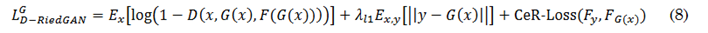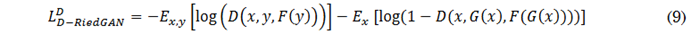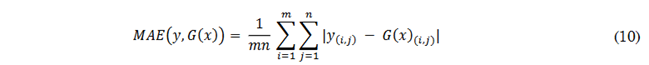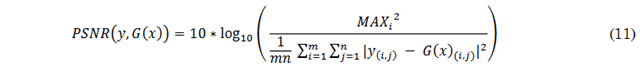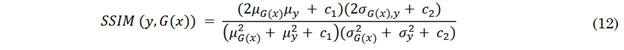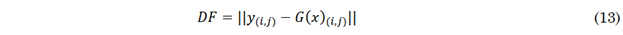1. INTRODUCTION
Breast cancer is a chronic non-communicable disease caused by DNA alterations that affect the normal division and growth of tissue cells. Due to its high incidence rates-particularly among the female population-it is one of the major public health concerns worldwide [1]. Moreover, it is a leading cause of cancer-related deaths globally [2]. According to the Global Cancer Observatory (GLOBOCAN), breast cancer accounted for 11.5 % of new cancer cases around the world in 2022 [3].
In general terms, breast cancer can be classified into five main types [4]. The first type, Lobular Carcinoma in Situ (LCIS), is a benign condition occurring in the breast lobules that does not spread outside of them. The second type, Ductal Carcinoma in Situ (DCIS), is a noninvasive neoplasm that develops in the milk glands or ducts without spreading beyond them. The third type, Infiltrating Ductal Carcinoma (IDC), begins in the ducts and may spread to surrounding breast tissues. The fourth type, Infiltrating Lobular Carcinoma (ILC), originates in the breast lobules and is often more challenging to detect due to its tendency to spread in a scattered manner rather than forming a lump. The fifth type, inflammatory breast cancer, is a rare and aggressive form that typically presents as redness, swelling, and warmth in the breast rather than a lump. It requires immediate treatment due to its rapid spread.
The treatment and prognosis of breast cancer depend on its type and specific characteristics. Nevertheless, early detection and timely treatment are crucial for preventing complications, improving patient prognosis, and reducing mortality rates [5]. Among the most commonly used methods for breast cancer detection are mammography and breast ultrasound. Mammography involves using low-dose X-rays to visualize internal breast tissues and is considered the standard detection method due to its low cost and ease of use [6]. However, it is only recommended for patients over 40 years of age and is contraindicated in pregnant women or in follow-ups due to radiation exposure. In contrast, ultrasound employs sound waves to visualize breast tissue [7], but it is highly dependent on the operator’s expertise. Furthermore, both methods may be ineffective in patients with dense breast tissue [6], [8].
When mammography or ultrasound results are inconclusive, more specialized tests are employed, which involve the intravenous administration of a contrast agent [9]. This chemical compound highlights cancer cells in the breast tissue by leveraging their elevated metabolic activity. Three such tests are Contrast-Enhanced Digital Mammography (CEDM), Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE-MRI), and Contrast-Enhanced Ultrasound (CEUS). CEDM requires taking two X-ray images of the breast: one before the administration of the contrast agent, employing low radiation levels (conventional mammography); and another after the application of the contrast agent, using high radiation. The two images are subsequently combined to create a recombined image, which highlights regions where the contrast agent has been absorbed in the tissue [10].
For its part, DCE-MRI uses magnetic waves to capture the absorption of the contrast agent over time, as it reacts in an accelerated manner in tissues with potential lesions [11]. To this end, the system takes a series of initial images of both breasts before the contrast agent is administered, followed by a series of images after its administration. This method is more sensitive and provides more accurate information about the state of the breast and any possible lesions, thus enabling the characterization of potential tumors. Regarding CEUS, it employs an intravascular contrast agent that allows real-time assessment of microcirculation and vascular and tissue perfusion [12].
Despite their advantages, these methods have limitations that include elevated costs, long acquisition times, and limited availability of equipment. Additionally, the contrast agent may cause allergic or adverse reactions in patients [13]. In the search for solutions to these setbacks, numerous studies have demonstrated the potential of deep learning neural networks to generate synthetic images that replicate the effect of the contrast agent without actually using it [14]-[18], while maintaining the visual quality of the images. On this background, the present study proposes a novel cost function, called Contrast-enhanced Region Loss (CeR-Loss), which leverages the contrast agent uptake behavior to generate synthetic post-contrast images from pre-contrast images in DCE-MRI studies. This function is employed in two new deep learning architectures, G-RiedGAN and D-RiedGAN, which focus on contrast-enhanced regions to improve the generation of synthetic post-contrast images.
2. LITERATURE REVIEW
Deep learning is a branch of machine learning that relies on artificial networks, which consist of interconnected layers of artificial neurons that can self-adjust based on the input and the amount of data they process [19]. One of the application areas of deep learning networks is image synthesis, which involves generating artificial images from a visual or textual description of their content. In the medical field, image synthesis has been employed, for example, for augmenting datasets to train models for disease diagnosis [20]; improving image resolution in specific imaging modalities [21]; segmenting regions of interest within images [22]; and generating images from data obtained through another examination modality [23]. The importance of image synthesis in medicine lies in its potential to enhance diagnostic accuracy, reduce the time and cost of capturing diagnostic images, and expand the availability of these medical examinations [24].
Image synthesis models can be broadly classified into two categories: autoencoders and Generative Adversarial Networks (GANs). Autoencoders, on the one hand, are architectures that comprise an encoder, which reduces the dimensionality of input data to learn an abstract (or latent) representation of its distribution; and a decoder, which reconstructs information from the latent space into a higher-dimensional space [25]. A variation of autoencoders, known as U-Net, addresses the information loss problem by copying information from the encoder layers to the decoder layers, thus improving the reconstruction of information in the higher-dimensional space [25], [26].
GANs, on the other hand, consist of a generator and a discriminator. The generator is a convolutional network that attempts to learn the latent distribution of the real data to generate synthetic information from a random noise sample. For its part, the discriminator is a complementary convolutional network that acts as an expert in distinguishing between real and synthetic information. The training of both networks is adversarial, that is, the generator strives to enhance its generation process to deceive the discriminator, while the discriminator seeks to refine its expertise to avoid being deceived by the generator. This adversarial learning process gives GANs their name [27].
Image synthesis methods can be employed to generate post-contrast images from pre-contrast images in DCE-MRI and CEDM studies. This application, known as domain shift, entails transforming a pre-contrast image (x) into a post-contrast image (y) [21], [23]. In this line, the authors of [15] trained a shallow generative architecture called SD-CNN to generate synthetic patches of post-contrast recombined images in CEDM studies from patches of full-field digital mammography images. In this study, two independent image repositories were used to extract the patches. The first dataset is INbreast [28], a public database from which 89 studies with BI-RADS categories 1 and 2 (benign) and 5 and 6 (malignant) were taken. The second database is proprietary and contains 49 studies with BI-RADS categories 4 and 5. All results were confirmed by biopsy, resulting in 23 benign and 26 malignant cases. The authors noted that using synthetic patches generated with their architecture improved the accuracy of mammography patch classifiers. However, a limitation of this architecture is that it is only capable of generating 3x3 synthetic patches from 15x15-pixel patches, which restricts its use in full image synthesis. Moreover, the shallowness of the network leads to few levels of abstraction, thus reducing its ability to synthesize complex structures such as those defining breast tissues.
In a subsequent study, the authors proposed a U-Net architecture called RiedNet [29]. This architecture introduced several modifications, including replacing pooling layers with convolutional and deconvolutional layers and incorporating a residual inception block to address the gradient fading problem caused by the network’s depth. The purpose of RiedNet was the synthetic generation of images in the medical context and its evaluation involved obtaining post-contrast recombined images of CEDM studies from low-energy images. Nevertheless, since the network was trained to synthesize 128x128-pixel blocks, the complete image synthesis considered the average of the generated blocks, potentially resulting in a blurring effect on the reconstructed breast tissues. In this case, the experiments were conducted on 139 contrasted mammography studies, with 112 used for training and 27 for testing.
Regarding the use of DCE-MRI studies for breast cancer detection, the authors of [30] employed a conditional GAN architecture called Pix2Pix [31], designed to generate fat-suppressed T1-weighted contrast-enhanced images from non-contrast images. The Pix2Pix architecture comprised a U-Net generator and a PatchGAN discriminator [31]. Particularly, the images used in this study were captured with a resolution of 3T and subsequently resized to 512x512 pixels. A total of 2,620 image pairs from 48 DCE-MRI studies were employed, with 2112 reserved for training, 418 for validation, and 90 for testing. Although the model tends to present errors in dense breast images, the authors emphasize the potential of the Pix2Pix architecture for synthetic generation of contrast-enhanced DCE-MRI images.
Another contribution to this field is the study presented in [17], which proposed a GAN architecture called TSGAN. TSGAN consists of four models: a U-Net model trained to generate post-contrast T1-weighted images from pre-contrast images; a global discriminator that focuses on differentiating real from fake post-contrast images; a local discriminator that distinguishes between real and fake regions of interest; and a U-Net model trained to generate segmentation masks over breast lesions. Similarly, in [32], the authors introduced an architecture called EDLS to synthesize dynamic sequences from T1WI images in MRI studies, improving lesion identification without using a contrast agent. Likewise, the authors of [33] explored the use of GAN architectures to generate realistic breast MRI images to enhance breast lesion detection. Additionally, in [34], the authors employed a CycleGAN architecture to translate images between different domains without the need for matched data pairs, thereby raising the quality of the synthesized images.
Furthermore, in [35], the authors presented a TDM-StarGAN architecture designed to generate synthetic images of conventional DCE-MRI study phases from ultrafast DCE-MRI study images. To this end, the authors modified the StarGAN architecture [29] for use with paired images. In addition, they considered the loss in the difference maps of the generated images and the detection area, obtained from the difference between the last post-contrast image and the pre-contrast image. They concluded that the proposed model outperformed the baseline models (Pix2Pix and StarGAN) by accurately synthesizing the regions associated with lesions.
In other imaging modalities, the study presented in [36] proposed the use of a Pix2Pix architecture to enhance the quality of low-count Positron Emission Tomography (dbPET) images, which is often compromised by patient respiration. Experiments were conducted on 49 cases, including 32 with abnormal results and 17 with normal results. To this end, each image was resized to 958x940 pixels. The authors posit that the Pix2Pix architecture can effectively address this problem by improving the quality of dbPET images with short acquisition times.
Aiming at reducing the radiation doses used in breast cancer diagnostic tests, the authors of [18] and [37] trained various models to generate digital mammography images from tomosynthesis images. The authors introduced an architecture called GGGAN, which employs a U-Net generator and a variant of the Pix2PixHD discriminator. The loss function for GGGAN uses the difference maps of the gradients of the images generated in specific intermediate layers.
In a context other than breast cancer diagnosis, the authors of [38] designed an architecture based on Pix2Pix, called Ea-GAN. It included the difference maps of the edges of the generated and real images in the loss function of both the discriminator and the generator. This inclusion aimed to enhance the synthesis of these elements and mitigate the smoothing effect commonly observed in U-Net networks [14]. This study employed the BRATS2015 database [39], which contains MRI images of 74 patients with a resolution of 240x240x155 voxels. The images were preprocessed and normalized with intensity values ranging from -1 to 1.
Despite numerous attempts to develop generative models for synthesizing diagnostic images in breast cancer detection, there are still significant limitations. This is largely due to the high variability in breast tissue density, which affects the performance of generative models when using contrast agents, as the visibility of these agents diminishes with increasing pixel intensity.
To address these challenges, this study proposes a novel architecture called D-RiedGAN. This architecture builds upon the Pix2Pix framework by incorporating residual inception blocks but focuses on contrast-enhanced regions in DCE-MRI studies.
3. METHODOLOGY
The proposed methodology begins with the implementation of a three-model baseline to synthesize fat-saturated T1-weighted images showing early response to contrast medium in DCE-MRI studies. Building on this baseline, two mixed architectures and two new architectures (G-RiedGAN and D-RiedGAN) are developed. These generative models are trained to generate synthetic post-contrast images, 𝑦̂ = 𝐺(𝑥). from non-contrast images, x. The goal is for the generative model, 𝐺(𝑥), to learn the early response to the contrast medium, thereby making the synthetic images resemble the real post-contrast images (y).
3.1 Optimization of contrast-enhanced regions
Traditional models for image synthesis have achieved significant advancements in natural image processing but face multiple limitations, especially when applied to specialized images such as medical ones. To overcome these challenges, this study proposes integrating a cost function that captures information from contrast-enhanced regions during training. This function aims to guide the synthesis process to accurately generate contrast enhancement in post-contrast images.
In terms of pixel intensity, the enhancement after contrast agent administration is identified by the highest intensities in the post-contrast image. Particularly, a global thresholding strategy, as shown in (1), is used to detect these high-intensity pixels. Here, y(i, j) is the pixel at position (i, j) in the post-contrast image and T is the threshold value.
Due to the sensitivity of T to intensity variations in images from different DCE-MRI studies, this parameter is set for each image using the 90th percentile of its histogram. In other words, the 10 % of the image pixels with the highest intensities are retained as contrast-enhanced regions. To refine these regions, closing and opening operators are employed using a 7x7-pixel circular structuring element, which smooths contours and removes small gaps between adjacent regions. This process is applied to both synthetic and real post-contrast images, generating real F y and synthetic F G(x) contrast enhancement masks.
Once the contrast-enhanced regions are identified, a cost function is used to minimize the discrepancies between these regions in real and synthetic images. Since contrast-enhanced regions are binary, optimizing them involves employing a cost function based on set similarity, such as the Jaccard index [40]. The Jaccard index between the real F y and generated (F G(x) ) contrast regions is computed using (2).
Because the Jaccard index is neither convex nor differentiable, its optimization via gradient descent (in the context of neural networks) can result in suboptimal solutions or convergence problems. [41] recommend employing a convex approximation to derive an optimizable function from the discrete function, which can then be optimized by first-order methods like gradient descent. This approach, based on the Lovász surrogate approximation, is estimated from a set of erroneous predictions, 𝑚(𝑐) ∈ 𝑅𝑝 for class 𝑐 ∈ 𝐶 rather than the vector of discrete predictions{0, 1}𝑝. Given that function ∆:{0, 1}𝑝 ↦ 𝑅𝑝 is submodular (como la función 𝐽𝑐), its Lovász extension is defined by (3).
Here, 𝑔𝑖(𝑚) = ∆({𝜋1. . . 𝜋𝑖 }) − ∆({𝜋1. . . 𝜋𝑖−1}) with 𝜋 being a permutation of the components of 𝑚 in descending order. Function ∆̅ is the strict convex closure of ∆, is piecewise linear, and interpolates the values of ∆, in 𝑅𝑝. Equation (4) is employed to compute the Lovász function of the Jaccard index in (2) (∆̅ 𝐽𝑐).
Where𝑓(𝑦, 𝑦∗) estimates the error vector m from the real or generated contrast masks after applying the SoftMax function. To avoid variations caused by batch size and the number of classes, the Lovász function is optimized by combining it with the Binary Cross-Entropy (BCE) described in (5), as suggested by the authors of [41].
Finally, to optimize the proposed models, a cost function combining BCE with the Lovász surrogate extension is used specifically for the contrast-enhanced regions. This combined function, termed CeR-Loss, is presented in (6).
3.2 G-RiedGAN y D-RiedGAN
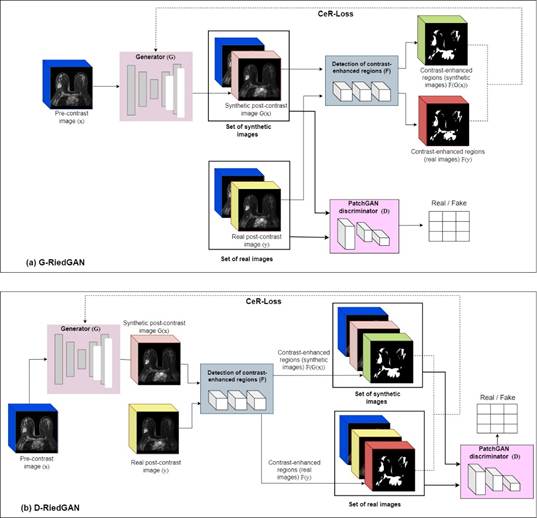
Figure 1a illustrates the general architecture of the first proposed model, G-RiedGAN. This architecture integrates, after the generator, a filter for detecting contrast-enhanced regions, which provides feedback to the generator to guide it in accurately replicating contrast enhancement. In this case, the PatchGAN discriminator, which identifies whether pre-contrast and post-contrast image pairs are real or synthetic, remains unchanged. The loss function of the G-RiedGAN generator, shown in (7), considers the overall loss from the pixel-level difference between the real and generated images, as well as the loss from the contrast-enhanced regions (CeR-Loss).
The proposed D-RiedGAN architecture, for its part, depicted in Figure 1b, includes the difference between the contrast-enhanced regions in both the generator and the discriminator. This allows the generator to focus more on these regions by considering them in the adversarial counterpart, thus improving the quality of the synthesized images.
For adversarial learning, the D-RiedGAN discriminator is modified to receive a triplet of images: the input image, the synthetic or real image, and the contrast-enhanced regions of the real or synthetic image. The loss functions of the D-RiedGAN generator and discriminator, defined in (8) and (9), respectively, include the loss from the contrast-enhanced regions (CeR-Loss).
3.3 Baseline models
The Pix2Pix [31], RiedNet [29], and Ea-GAN [38] architectures were employed as benchmarks to evaluate the proposed model. These architectures were chosen due to their proven effectiveness in tackling problems related to medical image synthesis across various modalities.
3.3.1 Pix2Pix
The Pix2Pix architecture [31] uses conditional information to guide the image generation process, allowing it to create an image in one domain based on an input image from another domain. It features a U-Net generator, a PatchGAN discriminator, and an objective function that combines adversarial loss (to make the generated images indistinguishable from the real ones) with pixel-level loss (to ensure content coherence between the generated and real images). A notable advantage of this architecture, as emphasized in the literature, is its capability to preserve fine details in the generated images, which is crucial for post-contrast image generation.
3.3.2 RiedNet
RiedNet [29] is an adapted U-Net architecture that employs convolutional and deconvolutional layers, along with a residual inception block to mitigate gradient fading issues. In this study, the RiedNet architecture was trained to synthesize complete images. Furthermore, the ReLU activation function in the intermediate layers was replaced with the Leaky ReLU function to maintain a small positive slope and avoid complete suppression of information in certain parts of the neural network [42]. The activation function of the output layer was also changed to the hyperbolic tangent function. These adjustments were made to preserve the value range of the input images during encoding.
3.3.3 Ea-GAN
Unlike the previous two architectures, Ea-GAN [38] incorporates edge information from both the original and synthesized images, calculated using a Sobel filter, into the learning process. This edge information helps the architecture focus on synthesizing the textures and edges of objects in the images. The Ea-GAN architecture has two variations: gEa-GAN, which integrates edge differences only in the generator’s loss function, and dEa-GAN, which includes edge differences in both the generator and discriminator.
To improve the synthesis process, these three foundational architectures were combined. The first combination, called RiedGAN, integrates a PatchGAN discriminator into the RiedNet architecture to enhance the synthesis process using an adversarial learning scheme. The key distinction between this network and the original Pix2Pix is that it employs the U-Net generator from the RiedNet architecture rather than the traditional U-Net generator.
Building on the idea of using edge maps from the Ea-GAN architecture, edge maps were incorporated into the RiedGAN framework, resulting in two new models: gEa-RiedGAN, which integrates edge maps into the RiedGAN generator, and dEa-RiedGAN, which incorporates edge map information into both the generator and the discriminator.
3.4 Evaluation metrics
To assess the quality of the synthetic images, three widely used quantitative metrics were employed: Mean Absolute Error (MAE), Peak Signal-to-Noise Ratio (PSNR), and Structural Similarity Index Measure (SSIM).
The Mean Absolute Error (MAE) quantifies the pixel-to-pixel difference between the intensities of two images. For a real image, y, and a generated image, 𝐺(𝑥), both of size 𝑚 𝑥 𝑛 pixels, MAE is computed as detailed in (10). A low MAE value signifies minimal error between the synthesized image and the reference image, with values close to 0 being ideal, i.e., indicating high accuracy. Conversely, a high MAE value reflects larger error and lower accuracy in image synthesis.
The Peak Signal-to-Noise Ratio (PSNR) represents the ratio between the maximum possible energy of a signal and the noise affecting the signal’s representation, measured in decibels (dB) [43]. The PSNR is defined by the formula in (11), where 𝑀𝐴𝑋𝑖 denotes the maximum possible intensity value for the images. A high PSNR value indicates greater similarity between the synthesized and reference images, while a low PSNR value suggests greater differences between them.
The Structural Similarity Index Measure (SSIM) considers the strong interdependencies between pixels, especially those in close proximity. These dependencies include information about luminance, contrast, and structure of the objects in the image and can be estimated jointly as shown in Equation (12) [43]. In this equation, 𝜇, 𝜎 and 𝜎2 represent the means, standard deviations, and covariances between the images, respectively, while ??1 and 𝑐2 are variables that stabilize the division in cases where the denominators are close to zero. An SSIM value close to 1 indicates high structural similarity between the synthesized image and the reference image, whereas a low SSIM value reflects reduced structural similarity. Values below 0.4 generally suggest poor quality in terms of image structure and texture.
Finally, difference maps are calculated by comparing individual pixels between a generated image and a real image to evaluate their discrepancies. This process is described by (13), where each pixel in the images is analyzed, and the difference in intensity between corresponding pixels in the two images is estimated. Each pixel’s value represents its intensity, and pixel comparison involves subtracting the value of the corresponding pixel in one image from the value of the same pixel in the other image. This comparison is used to quantify and visualize the differences between the generated and real images.
4. RESULTS AND DISCUSSION
4.1 Experimental setup
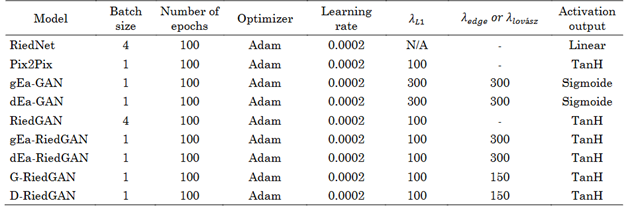
The results presented in this study were obtained using the experimental setup detailed in Table 1. This table outlines the hyperparameters employed for each model, which were adjusted according to the available computational resources. The experiments were conducted on a workstation equipped with an Intel Xeon Silver 4108 CPU and an NVIDIA Quadro P2000 GPU with 4 GB of RAM. Python version 3.8 was used as the programming language, along with PyTorch version 2.0.
4.2 Database
A proprietary, retrospective, and anonymized database of DCE-MRI studies from 197 patients was used to train the models. Each study includes T1- and T2-weighted structural images, Diffusion Weighted images (DWIs), and six DCE images. This study focused on the T1 fat-saturated sequence acquired before contrast agent administration x and the corresponding image acquired in the early stage after contrast agent administration y.
Given the retrospective nature of the database, studies were selected based on their use of different types of 1.5 T resonators and gadolinium-based contrast agents, with doses ranging between 0.014 and 0.016 ml/mol. All studies contained at least one abnormality (either benign or malignant) annotated by expert radiologists using the BI-RADS system. This selection process ensured a balanced representation of both benign and malignant cases.
To focus on synthesizing contrast regions, only images with annotated contrast regions were included to ensure accurate depiction of contrast uptake. As a result, a total of 937 normalized images, scaled to the range from -1 to 1, were processed. Of these, 718 images were allocated for training and 219 for validation. The original images, with resolutions ranging between 480x480 and 512x512 pixels, were resized to 240x240 pixels for consistency purposes.
4.3 Comparative evaluation
Figure 2 presents a comparison of the PSNR, SSIM, and MAE metrics for the models assessed on the validation image set. As observed, the G-RiedGAN and D-RiedGAN models proposed in this study outperformed the other models. This suggests that incorporating contrast-enhanced regions into the image synthesis process using the CeR-Loss function significantly enhances the quality of the synthetic images, as reflected by the values of these quantitative metrics.

Source: Own work.
Figure 2 Scatter plot comparing PSNR, SSIM, and MAE across the baseline, mixed, and proposed models
Although G-RiedGAN and D-RiedGAN showed a slightly higher MAE compared to RiedGAN, the increase in MAE for D-RiedGAN was minimal and is outweighed by substantial improvements in PSNR and SSIM. This indicates that while RiedGAN demonstrated slightly better accuracy in individual pixel errors, it tends to produce more blurred images of internal structures, making it less suitable for medical image synthesis.
In comparison to Pix2Pix [31]-a model frequently used in similar studies-both G-RiedGAN and D-RiedGAN exhibited superior performance in terms of PSNR and SSIM. They excelled particularly in synthesizing contrast-enhanced regions and reducing noise, thus addressing some of the limitations of Pix2Pix in dense breast imaging.
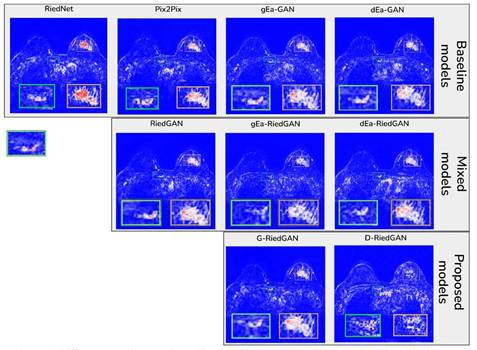
Figure 3, for its part, displays real post-contrast images generated from their non-contrast counterparts. Overall, the models effectively reproduced larger anatomical structures, though some discrepancies were observed in the intensities of rib cage structures. Despite this, G-RiedGAN and D-RiedGAN showed superior synthesis of contrast-enhanced regions compared to reference models such as RiedNet [29], Pix2Pix [31], and EaGAN [32]. These reference models, which served as the foundation for G-RiedGAN and D-RiedGAN, were employed for comparative analysis with the same methodology. While effective in their respective contexts, this study specifically evaluated their performance with medical images.

Source: Own work.
Figure 3 Results of the comparative analysis between the evaluated models. The zoomed regions highlight projections where the contrast agent was captured
Based on the results, G-RiedGAN and D-RiedGAN combine the strengths of existing models to achieve more precise and higher-quality image synthesis, particularly in contrast-enhanced regions. They also performed well when compared to models reported in the literature, notably in minimizing noise and blur. This is further illustrated in Figure 4, which shows the difference maps of the synthetic and real images. As can be seen, the difference maps for G-RiedGAN and D-RiedGAN reveal fewer discrepancies between the synthetic and real images.
4.4 Impact of the contrast-enhanced regions’ cost function (CeR-Loss)
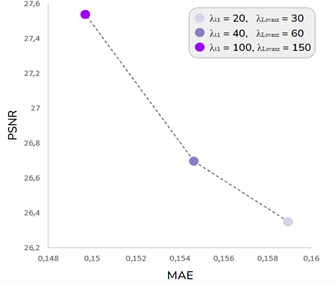
The proposed cost function, CeR-Loss, is a critical component of the D-RiedGAN architecture, significantly enhancing its performance when compared to other models. To evaluate the impact of this function on model training, a series of experiments were conducted using the D-RiedGAN architecture (described in the previous section) but with variations in parameters 𝜆𝑙1 and 𝜆 𝐿𝑜𝑣𝑎𝑠𝑧 . In these experiments, 𝜆 𝐿𝑜𝑣𝑎𝑠𝑧 was consistently set higher than 𝜆 𝑙1 across all configurations.
Figure 5 illustrates the results for three different parameter settings: 𝜆 𝑙1 = 20 and 𝜆 𝐿𝑜𝑣𝑎𝑠𝑧 = 30; 𝜆𝑙1 = 40 and 𝜆 𝐿𝑜𝑣𝑎𝑠𝑧 = 60; and 𝜆𝑙1 = 100 and 𝜆 ??𝑜𝑣𝑎𝑠𝑧 = 150. As observed, both the MAE and PSNR metrics improved as the values of 𝜆𝑙1 and 𝜆 𝐿𝑜𝑣𝑎𝑠𝑧 was increased, with the best performance achieved at 𝜆 𝑙1 = 100 and 𝜆 𝐿𝑜𝑣𝑎𝑠𝑧 = 150. These results underscore the positive effect of the CeR-Loss cost function on the model’s overall performance.
5. CONCLUSIONS
This paper introduced CeR-Loss, a novel cost function designed to leverage contrast agent uptake for generating synthetic post-contrast images from pre-contrast images in DCE-MRI studies. This function is incorporated into two new deep learning architectures, G-RiedGAN and D-RiedGAN, which focus on contrast-enhanced regions to improve the synthesis of post-contrast images. The primary goal of these architectures is to minimize dependence on contrast agents and reduce the costs associated with DCE-MRI studies for breast cancer screening.
The proposed G-RiedGAN and D-RiedGAN models combine features from the RIED-Net and Pix2Pix architectures within the EaGAN framework. Notably, D-RiedGAN includes a filter for detecting contrast-enhanced regions, which are essential for accurately synthesizing DCE-MRI images in breast cancer detection and diagnosis. These identified contrast regions guide the network learning process through the Lovász and BCE loss functions, which are integrated into the loss function of the generator and the discriminator (CeR-Loss).
Two approaches were used for comparative evaluation. The first approach compared the performance of the proposed models (with CeR-Loss) against models documented in the literature and a set of mixed models. The results, based on MAE, PSNR, and SSIM metrics, indicate that the proposed models more effectively synthesize contrast-enhanced regions, with reduced noise and blur. The second approach assessed the impact of the CeR-Loss function on the learning process, revealing that increasing the weight of CeR-Loss positively influences the synthesis of contrast regions, as reflected by the value of the same metrics.
Although validation was limited to quantitative metrics based on pixel intensities of synthetic images, future studies should include qualitative assessments by expert radiologists to validate the diagnostic quality of these images. Additionally, future research should investigate the performance of the baseline and proposed models across heterogeneous image databases, considering variations in study quality (0.5 T, 1.5 T, 3 T, and 7 T), dosage, and contrast agents. Furthermore, incorporating synthetic post-contrast images could improve the training of breast cancer detection and classification models that use conventional MRI images, as these synthetic images might provide valuable supplementary information to enhance model performance.











 text in
text in